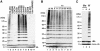A ubiquitin ligase complex assembles linear polyubiquitin chains - PubMed (original) (raw)
A ubiquitin ligase complex assembles linear polyubiquitin chains
Takayoshi Kirisako et al. EMBO J. 2006.
Abstract
The ubiquitin system plays important roles in the regulation of numerous cellular processes by conjugating ubiquitin to target proteins. In most cases, conjugation of polyubiquitin to target proteins regulates their function. In the polyubiquitin chains reported to date, ubiquitin monomers are linked via isopeptide bonds between an internal Lys and a C-terminal Gly. Here, we report that a protein complex consisting of two RING finger proteins, HOIL-1L and HOIP, exhibits ubiquitin polymerization activity by recognizing ubiquitin moieties of proteins. The polyubiquitin chain generated by the complex is not formed by Lys linkages, but by linkages between the C- and N-termini of ubiquitin, indicating that the ligase complex possesses a unique feature to assemble a novel head-to-tail linear polyubiquitin chain. Moreover, the complex regulates the stability of Ub-GFP (a GFP fusion protein with an N-terminal ubiquitin). The linear polyubiquitin chain generated post-translationally may function as a new modulator of proteins.
Figures
Figure 1
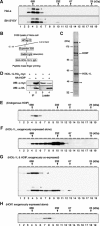
Identification of HOIP, a component of the 600 K complex. (A) HOIL-1L forms an ∼600 kDa complex in HeLa and SH-SY5Y cells. Fractions from S100 lysates separated by Superdex 200 HR were subjected to SDS–PAGE, followed by immunoblotting with anti-HOIL-1. * indicates an unidentified modified form of HOIL-1L. (B) Procedure for isolation of the 600 K complex and identification of HOIP. Details are described in Materials and methods. (C) Coomassie staining of the proteins in the final fraction collected using the procedure outlined in (B). * represents a protein which crossreacted with the antibody used for purification and ** is actin. (D) HOIP binds to HOIL-1L. Anti-HA immunoprecipitates (IP) and cell lysates from U2OS cells expressing HOIL-1L-His6-myc alone (lanes 1 and 3) or those of U2OS cells co-expressing HOIL-1L-His6-myc and HA-HOIP (lanes 2 and 4) were separated by SDS–PAGE, followed by immunoblotting with rabbit anti-myc (α-myc) or mouse anti-HA (α-HA). (E) HOIP has a distribution similar to HOIL-1L. Fractionated S100 lysates of HeLa cells were probed with anti-HOIP. (F) HOIL-1L-His6-myc forms the 600 K complex poorly when expressed alone in HeLa cells. Fractions from S100 lysates of HeLa cells expressing HOIL-1L-His6-myc were probed by immunoblotting using anti-myc. (G) Co-expression of HA-HOIP (arrowhead) and HOIL-1L-His6-myc (arrow) is sufficient for formation of the 600 K complex in HeLa cells. Fractionated S100 lysates of HeLa cells expressing both HOIL-1L-His6-myc and HA-HOIP were probed with anti-HA or anti-myc. * indicates an unidentified modified form of HOIL-1L. (H) HA-HOIP forms the 600 K complex poorly when expressed alone in HeLa cells. Fractionated S100 lysates of HeLa cells expressing HA-HOIP were probed with anti-HA.
Figure 2
Specific interaction between the UBA of HOIP and the UBL of HOIL-1L is crucial for the 600 K complex formation. (A) Schematic representation of HOIP and HOIL-1L. ZF, zinc-finger domain; NZF, Npl4 type ZF domain; UBA, ubiquitin associated domain; UBL, ubiquitin like domain; IBR, in between ring domain; and RBR, RING-IBR-RING domain. (B) The UBL of HOIL-1L is required for the binding between HOIL-1L and HOIP. Anti-Flag immunoprecipitates (IP) and cell lysates from U2OS cells co-expressing Flag-HOIP and C-terminally HA-tagged HOIL-1L (1), or co-expressing Flag-HOIP and the HA-tagged HOIL-1L domain-deleted mutants (2, 3 or 4) were separated by SDS–PAGE and subjected to immunoblotting with anti-HA (α-HA) or anti-Flag (α-Flag). * represents a nonspecific signal. (C) The UBA of HOIP is required for binding HOIL-1L. Anti-Flag immunoprecipitates (IP) and cell lysates from U2OS cells co-expressing HOIL-1L-His6-Flag and myc-HOIP (1) or co-expressing HOIL-1L-His6-Flag and myc-HOIP domain-deleted mutants (2, 3 or 4) were separated by SDS–PAGE, followed by immunoblotting with anti-myc (α-myc) or anti-Flag (α-Flag). (D) The UBA of HOIP directly recognizes the UBL of HOIL-1L. Mixtures of bacterially purified His6-Xpress-tagged HOIP UBA (aa 556–636) and GST-HOIL-1L UBL (aa 1–130) (2), GST-ubiquitin (3), GST (4), and GST-HOIL-1L UBL (aa 1–130) alone (1) were incubated with Ni2+-affinity gel. Proteins bound to the Ni2+-affinity gel and 5% input samples were separated by SDS–PAGE and subjected to immunoblotting with anti-GST (α-GST) or anti-Xpress (α-Xpress).
Figure 3
The 600 K complex assembles polyubiquitin chains in vitro. (A) The 600 K complex from insect cells (Complex) ubiquitinates Ub-GST. Ub-GST or GST were incubated with ubiquitin and ATP, together with the indicated components at 37°C for 2 h, followed by immunoblotting with anti-GST. Asterisks indicate nonspecific signals. (B) The 600 K complex ubiquitinates Ub-GFP. Ub-GFP or GFP were incubated as described in (A), followed by immunoblotting with anti-GFP. (C) The 600 K complex polymerizes ubiquitin. Ubiquitin was incubated, as indicated, together with ATP at 37°C for 2 h, followed by immunoblotting with anti-ubiquitin. (D) The 600 K complex exhibits much greater ubiquitination activity than either HOIL-1L or HOIP alone. Ubiquitin was incubated as in (C) together with HOIL-1L (lane 1), HOIP (lane 2) or the 600 K complex (lane 3), followed (by immunoblotting with) anti-ubiquitin, anti-HOIP or anti-HOIL-1. (E) RING finger domains of HOIP but not those of HOIL-1L are critical for the ubiquitin conjugation activity of the 600 K complex. Ubiquitin was incubated as in (C) in the presence of the WT 600 K complex, or the mutant 600 K complex containing a RING finger mutant of either HOIL-1L or HOIP, as indicated, at 37°C for 2 h, followed by immunoblotting as described in (D). (F) The 600 K complex conjugates only one methylated ubiquitin to Ub-GST. Ub-GST was incubated with methylated ubiquitin (Me) at 4°C (lane 1) or 37°C (lane 2), or with WT ubiquitin (W) at 37°C (lane 3) in the presence of E1, UbcH5C, ATP and the 600 K complex from insect cells, followed by immunoblotting as described in (A). The arrow and * indicate mono-ubiquitinated Ub-GST and a nonspecific signal, respectively.
Figure 4
The 600 K complex assembled the N-terminal Met-linked polyubiquitin chain with a broad spectrum of E2s. (A) E2-25K, HHR6B, UbcH5s and UbcH7 could function as E2s for the 600 K complex to generate polyubiquitin chains. Ubiquitin was incubated with the indicated E2s in the presence of E1, the 600 K complex and ATP, followed by immunoblotting with anti-ubiquitin. (B) The 600 K complex polymerized Lys-less and K1 ubiquitin. WT (lane 1), the Lys-less (K0, lane 2) and the K1 ubiquitin mutants (six out of seven Lys are mutated to arginines and the number represents the remaining Lys residue) (lanes 3–9) were incubated with E1, UbcH5C, the 600 K complex and ATP, followed by immunoblotting as described in (A). (C) The 600 K complex failed to polymerize methylated ubiquitin. Methylated (Me) and WT ubiquitin (W) were incubated as in (B) at 4°C (lane 1) or 37°C (lanes 2 and 3), followed by immunoblotting as in (A). The arrow represents possible methylated di-ubiquitin contamination in the methylated ubiquitin preparation.
Figure 5
Mass spectrometric analyses of the N-terminally-linked head-to-tail polyubiquitin chains generated by the 600 K complex. (A) Coomassie staining of polyubiquitin chains. Ubiquitin was incubated with E1, ATP, the indicated E2s in the presence (lanes 2–5) or absence (lane 1) of the 600 K complex at 37°C for 2 h. Ubiquitin was also incubated with E1, ATP, non-tagged E2-25K in the absence of the 600 K complex at 37°C for 12 h (lane 6) or in the presence of the 600 K complex at 37°C for 2 h (lane 7). The reactions were stopped by heating at 60°C for 15 min, followed by centrifugation at 20 000 g for 20 min. The resulting supernatants were separated by SDS–PAGE and stained with Coomassie. *, ** and *** indicate His6-E2-25K, His6-UbcH7 and non-tagged E2-25K, respectively. (B) Mass spectrometry analysis of the polyubiquitin chains generated by the 600 K complex and UbcH5C. 1–2 × Ub and the polyubiquitin chain mixtures (polyUb) generated by UbcH5C and the 600 K complex as depicted in (A, lane 4) were digested with trypsin in the gels at 37°C for 12 h, followed by MALDI-TOF/MS. (M1) indicates the fragment specifically generated from the N-terminally linked ubiquitin chain. (a–g) represent the ubiquitin fragments of amino acids 43–48, 1–6, 34–42, 64–72, 55–63, 30–42 and 12–27, respectively. (C) MS/MS spectrum acquired from the M1 peptide (m/z+=879.48–879.58). The spectrum was obtained by the Lift analysis according to the manufacturer's instructions using ultraflex TOF/TOF and interpreted by the Mascot Search Program. The amino-acid sequences from the C- and N-terminus (the starting point is on the left) were shown on the top based on y m and b_i_ (where m and l denote positions counted from C- and N-terminus) that were produced by cleavage of peptide bonds during MS/MS. (D, E) Mass spectrometry analysis of polyubiquitin chains generated by E2-25K in the absence (D) or presence (E) of the 600 K complex. 2 × Ub and polyUb generated by E2-25K in the presence or absence of the 600 K complex depicted in (A, lanes 6 and 7) were subjected to MALDI-TOF/MS as described in (B). (K48) indicates the fragment specifically generated from the K48-linked ubiquitin chain. (M1) and (a–g) are the same as (B).
Figure 6
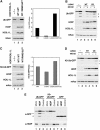
In vivo roles of LUBAC. (A) LUBAC induces the decrease of Ub-GFP. HeLa cells were co-transfected with expression plasmids encoding Ub-GFP together with mitochondrial aconitase-myc (mAco: transfection control), HOIL-1L-WT and HOIP-WT or HOIP-RING1&2CS as indicated. Total cell lysates were probed with anti-GFP (Ub-GFP), anti-FLAG (HOIP) anti-HA (HOIL-1L) or anti-myc (mAco). Relative amounts of Ub-GFP shown in the bar graph were quantified by LAS3000. (B) LUBAC enhances the proteasomal degradation of Ub-GFP. HeLa cells were co-transfected with expression plasmids encoding Ub-GFP and mAco together with control plasmids (vec: lanes 1 and 2), HOIL-1L-WT and HOIP-WT (WT: lanes 3 and 4) or HOIL-1L-WT and HOIP-RING1&2CS (CS: lanes 5 and 6). MG132 was added for 8 h before harvesting cells (lanes 2, 4 and 6). Total cell lysates were probed as in (A). (C) LUBAC induces a decrease in K0-Ub-GFP. HeLa cells were co-transfected as indicated in (A), with the exception that the expression plasmid for K0-Ub-GFP was transfected instead of the Ub-GFP plasmid. Total cell lysates were probed as in (A). (D) LUBAC enhances the proteasomal degradation of K0-Ub-GFP. HeLa cells were co-transfected as indicated in (B) except that the expression plasmid for K0-Ub-GFP was transfected instead of the Ub-GFP plasmid. Total cell lysates were probed as in (A). (E) Knockdown of HOIP increased the amount of Ub-GFP and K0-Ub-GFP. HeLa cells were transfected with si-RNA for HOIP (lane 2, 5 and 8), scrambled contol siRNA (lanes 3, 6 and 9) or mock (lanes 1, 4 and 7) followed by transfection of expression plasmids encoding Ub-GFP (lanes 1–3), K0-Ub-GFP (lanes 4–6) or GFP (lanes 7–9). Total cell lysates were probed with anti-GFP (Ub-GFP, K0-Ub-GFP or GFP) or anti-HOIP. Asterisks indicate possible degradation products of (K0-)Ub-GFP.
Similar articles
- Solution structure of the HOIL-1L NZF domain reveals a conformational switch regulating linear ubiquitin affinity.
Walinda E, Sugase K, Ishii N, Shirakawa M, Iwai K, Morimoto D. Walinda E, et al. J Biol Chem. 2023 Sep;299(9):105165. doi: 10.1016/j.jbc.2023.105165. Epub 2023 Aug 16. J Biol Chem. 2023. PMID: 37595872 Free PMC article. - Functions of Linear Ubiquitin Chains in the NF-κB Pathway : Linear Polyubiquitin in NF-κB Signaling.
Iwai K. Iwai K. Subcell Biochem. 2010;54:100-6. doi: 10.1007/978-1-4419-6676-6_8. Subcell Biochem. 2010. PMID: 21222276 - The zinc finger of the CSN-associated deubiquitinating enzyme USP15 is essential to rescue the E3 ligase Rbx1.
Hetfeld BK, Helfrich A, Kapelari B, Scheel H, Hofmann K, Guterman A, Glickman M, Schade R, Kloetzel PM, Dubiel W. Hetfeld BK, et al. Curr Biol. 2005 Jul 12;15(13):1217-21. doi: 10.1016/j.cub.2005.05.059. Curr Biol. 2005. PMID: 16005295 - The emerging role of linear ubiquitination in cell signaling.
Emmerich CH, Schmukle AC, Walczak H. Emmerich CH, et al. Sci Signal. 2011 Dec 20;4(204):re5. doi: 10.1126/scisignal.2002187. Sci Signal. 2011. PMID: 22375051 Review. - Linear ubiquitin chains: enzymes, mechanisms and biology.
Rittinger K, Ikeda F. Rittinger K, et al. Open Biol. 2017 Apr;7(4):170026. doi: 10.1098/rsob.170026. Open Biol. 2017. PMID: 28446710 Free PMC article. Review.
Cited by
- Regulation of translesion DNA synthesis: Posttranslational modification of lysine residues in key proteins.
McIntyre J, Woodgate R. McIntyre J, et al. DNA Repair (Amst). 2015 May;29:166-79. doi: 10.1016/j.dnarep.2015.02.011. Epub 2015 Feb 18. DNA Repair (Amst). 2015. PMID: 25743599 Free PMC article. Review. - Parkin-catalyzed ubiquitin-ester transfer is triggered by PINK1-dependent phosphorylation.
Iguchi M, Kujuro Y, Okatsu K, Koyano F, Kosako H, Kimura M, Suzuki N, Uchiyama S, Tanaka K, Matsuda N. Iguchi M, et al. J Biol Chem. 2013 Jul 26;288(30):22019-32. doi: 10.1074/jbc.M113.467530. Epub 2013 Jun 10. J Biol Chem. 2013. PMID: 23754282 Free PMC article. - A MUB E2 structure reveals E1 selectivity between cognate ubiquitin E2s in eukaryotes.
Lu X, Malley KR, Brenner CC, Koroleva O, Korolev S, Downes BP. Lu X, et al. Nat Commun. 2016 Aug 23;7:12580. doi: 10.1038/ncomms12580. Nat Commun. 2016. PMID: 27550514 Free PMC article. - Regulation of RIPK1 Phosphorylation: Implications for Inflammation, Cell Death, and Therapeutic Interventions.
Du J, Wang Z. Du J, et al. Biomedicines. 2024 Jul 9;12(7):1525. doi: 10.3390/biomedicines12071525. Biomedicines. 2024. PMID: 39062098 Free PMC article. Review. - Specific recognition of linear ubiquitin chains by the Npl4 zinc finger (NZF) domain of the HOIL-1L subunit of the linear ubiquitin chain assembly complex.
Sato Y, Fujita H, Yoshikawa A, Yamashita M, Yamagata A, Kaiser SE, Iwai K, Fukai S. Sato Y, et al. Proc Natl Acad Sci U S A. 2011 Dec 20;108(51):20520-5. doi: 10.1073/pnas.1109088108. Epub 2011 Dec 2. Proc Natl Acad Sci U S A. 2011. PMID: 22139374 Free PMC article.
References
- Bayle J, Lopez S, Iwai K, Dubreuil P, De Sepulveda P (2006) The E3 ubiquitin ligase HOIL-1 induces the polyubiquitination and degradation of SOCS6 associated proteins. FEBS Lett 580: 2609–2614 - PubMed
- Ben-Saadon R, Fajerman I, Ziv T, Hellman U, Schwartz AL, Ciechanover A (2004) The tumor suppressor protein p16(INK4a) and the human papillomavirus oncoprotein-58 E7 are naturally occurring lysine-less proteins that are degraded by the ubiquitin system. Direct evidence for ubiquitination at the N-terminal residue. J Biol Chem 279: 41414–41421 - PubMed
- Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, Reed SI (2001) UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat Struct Biol 8: 417–422 - PubMed
- Bloom J, Amador V, Bartolini F, DeMartino G, Pagano M (2003) Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell 115: 71–82 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases