The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies - PubMed (original) (raw)
The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies
Marisa S Otegui et al. Plant Cell. 2006 Oct.
Abstract
We have investigated the transport of storage proteins, their processing proteases, and the Vacuolar Sorting Receptor-1/Epidermal Growth Factor Receptor-Like Protein1 (VSR-1/ATELP1) receptor during the formation of protein storage vacuoles in Arabidopsis thaliana embryos by means of high-pressure freezing/freeze substitution, electron tomography, immunolabeling techniques, and subcellular fractionation. The storage proteins and their processing proteases are segregated from each other within the Golgi cisternae and packaged into separate vesicles. The storage protein-containing vesicles but not the processing enzyme-containing vesicles carry the VSR-1/ATELP1 receptor. Both types of secretory vesicles appear to fuse into a type of prevacuolar multivesicular body (MVB). We have also determined that the proteolytic processing of the 2S albumins starts in the MVBs. We hypothesize that the compartmentalized processing of storage proteins in the MVBs may allow for the sequential activation of processing proteases as the MVB lumen gradually acidifies.
Figures
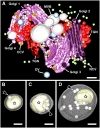
Figure 1.
Tomographic Models of Golgi Stacks, Associated Vesicles, and Secretory Compartments during PSV Formation in the Arabidopsis Embryo Reconstructed from a Serial Tomogram Consisting of Five Sections. (A) A cluster of four Golgi stacks delimits an area with high concentrations of TGNs, dense vesicles (DV), clathrin-coated vesicles (CCV), noncoated vesicles (NCV), and MVBs. Bar = 200 nm. (B) to (D) MVB compartments. The limiting membranes of MVBs have been made translucent in these tomographic models to visualize the internal vesicles (arrows) and electron-dense aggregates (stars). Bars = 100 nm.
Figure 2.
Golgi Stacks and Associated Compartments during the Formation of PSVs. (A) Tomographic slice (4.3 nm thick) through a Golgi stack and associated vesicles and MVBs. (B) Tomographic model derived from the tomogram depicted in (A). Based on structural and staining patterns, this Golgi stack appears to consist of two _cis_-cisternae, three medial cisternae, one _trans_-cisterna, and a partially detached TGN. (C) Face-on views of Golgi cisternae. Storage protein aggregates accumulate in the margins of all cisternae (arrows) except the _cis_-most cisterna. CC, clathrin coat. Bars = 100 nm.
Figure 3.
Distribution of Storage Proteins, Processing Proteases, and the VSR-1/ATELP1 Receptor in the Golgi Stacks and Dense Vesicles. (A) Immunolabeling of storage proteins with anti-2S albumin antibodies. Note the high concentration of gold particles over the marginal electron-dense aggregates of the Golgi cisternae (arrows). (B) and (C) Double immunolabeling with anti-2S albumin antibodies (5-nm gold particles) and anti-12S globulin antibodies (15-nm gold particles). (D) to (H) Double immunolabeling of marginal buds in Golgi cisternae ([D] and [E]) and dense vesicles ([F] to [H]) with anti-VSR-1/ATELP1 antibodies (5-nm gold particles) and anti-2S albumin antibodies (15-nm gold particles). (I) and (J) Double immunolabeling of Golgi stacks with anti-aspartic protease A1 antibodies (AtAP; 5-nm gold particles; arrows) and anti-2S albumin antibodies (15-nm gold particles; arrowheads). Bars = 100 nm.
Figure 4.
Immunolabeling of Golgi Stacks and PSVs in the Arabidopsis Embryo. (A) Golgi stack (G) and MVB labeled with anti-β-VPE antibodies (arrows). (B) PSV labeled with anti-VSR-1 antibodies (arrowheads). (C) PSV labeled with anti-aspartic protease A1 antibodies (AtAP; arrowheads). (D) PSV labeled with anti-β-VPE antibodies (arrowheads). Bars = 100 nm.
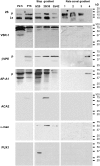
Figure 5.
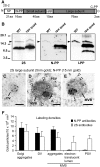
Protein Gel Blot Analysis of the Subcellular Fractions from B. napus Embryos. P2.5 and P16 are the pellet fractions collected after the 2,500_g_ and 16,000_g_ centrifugation steps, respectively. The 16,000_g_ supernatant was applied onto a sucrose density step gradient (20, 35, 42, and 55% [w/v] sucrose) and centrifuged at 85,500_g_ for 120 min. The 42 to 55% sucrose interphase, which is enriched in secretory vesicles (Hinz et al., 1999), was layered onto a linear sucrose density gradient (20 to 55% sucrose) and centrifuged at 16,000_g_ for 20 min (rate-zonal gradient). The rate-zonal gradient fractions 1 to 4 were collected from the top of the tube. Fractions 1 (22% sucrose) to 3 (28% sucrose) contain the 2S albumin precursors and the VSR-1/ATELP1 receptor but not the processing enzymes β-VPE and aspartic protease A1 (AtAP), whereas fraction 4 (31% sucrose) contains the processing enzyme precursors but not the storage proteins. Antibodies against subcellular markers were used to ensure the purity of the fractions: Arabidopsis Ca2+-ATPase2 (ACA2) for the ER, α-mannosidase (α-man) for the Golgi, and Arabidopsis UBX Domain–containing Protein1 (PUX1) for the cytoplasm. Ls, 2S albumin large subunit; p, precursor.
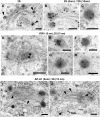
Figure 6.
Immunolabeling of MVBs during PSV Formation in Arabidopsis Embryo Cells. (A) to (C) Antibodies against storage proteins. Single immunolabeling of 2S albumins (A) and 12S globulins (B), and double immunolabeling of 2S albumins (15-nm gold particles) and 12S globulins (5-nm gold particles) (C). (D) and (E) Antibodies against proteolytic processing enzymes: anti-β-VPE (D) and anti-aspartic protease A1 (AtAP) (E) antibodies. (F) Double immunolabeling of aspartic protease A1 (5-nm gold particles) and 2S albumins (15-nm gold particles). (G) and (H) Antibodies against the VSR-1/ATELP1 receptor. (I) Double immunolabeling of the VSR-1/ATELP1 receptor (5-nm gold particles) and 2S albumins (15-nm gold particles). Bars = 100 nm.
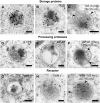
Figure 7.
Detection of 2S Albumin Propeptides. (A) Diagram of an Arabidopsis 2S albumin precursor: signal peptide (SP), three propeptides (N-terminal propeptide [N-PP], internal propeptide [I-PP], and C-terminal propeptide [C-PP]), and the small and large chains. aa, amino acids. (B) Protein gel blots of protein extracts from both wild-type and _vpe-_quadruple mutant Arabidopsis seeds. The anti-2S albumin antibodies (2S) recognize the 2S albumin large chain in both the precursor (∼15 kD) and the mature (∼7 kD) forms; the I-PP peptide antibodies recognize the 2S albumin internal propeptide; and the N-PP peptide antibodies recognize the 2S albumin N-terminal propeptide. Arrowheads indicate the 2S albumin precursors. (C) to (E) Double immunolabeling of a dense vesicle (C) and MVBs ([D] and [E]) with anti-2S albumin antibodies (5-nm gold particles) and anti-N-PP antibodies (15-nm gold particles). The less electron-dense luminal contents that surround the dense aggregate inside MVBs are labeled with anti-2S albumin antibodies (arrowheads) but are very scarcely labeled with anti-N-PP antibodies. Bars = 100 nm. (F) Labeling density on different organelles with the N-PP and 2S albumin antibodies. Values shown are means of 10 to 15 sampled areas, and error bars indicate
se
.
Figure 8.
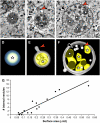
Formation of MVBs. (A) to (C) Tomographic slices of an electron-dense vesicle (A), a putative intermediate pre-MVB compartment (B), and an MVB (C) in Arabidopsis embryo cells. Bar = 100 nm. (D) to (F) Three-dimensional tomographic models corresponding to the structures depicted in (A) to (C), respectively. The electron-dense aggregates of storage proteins are indicated by stars. The putative pre-MVB depicted in (B) and (E) shows a vesicle-budding/fusing profile (arrowheads). The MVB depicted in (C) and (F) shows a forming internal vesicle invaginating from the limiting membrane (arrowheads) and numerous internal vesicles (arrows). (G) Relationship between the surface area of MVBs and the number of internal vesicles. Surface area and number of internal vesicles were calculated for 25 intermediate and MVB compartments using IMOD software.
Figure 9.
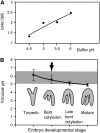
Determination of the Luminal pH of PSVs Using Lysosensor Yellow/Blue. (A) Calibration curve. (B) pH inside PSVs during embryo development. The arrow indicates the developmental stage at which storage proteins are first detected inside the PSVs by immunolabeling. The shaded area represents the pH range in which the solubility of the processed 12S globulins is <50%, as reported by Gruis et al. (2004).
Figure 10.
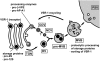
Model Depicting the Sorting and Trafficking of Storage Proteins and Processing Proteases in Arabidopsis Embryos during PSV Formation.
Similar articles
- Protein mobilization in germinating mung bean seeds involves vacuolar sorting receptors and multivesicular bodies.
Wang J, Li Y, Lo SW, Hillmer S, Sun SS, Robinson DG, Jiang L. Wang J, et al. Plant Physiol. 2007 Apr;143(4):1628-39. doi: 10.1104/pp.107.096263. Epub 2007 Feb 23. Plant Physiol. 2007. PMID: 17322331 Free PMC article. - Localization of vacuolar transport receptors and cargo proteins in the Golgi apparatus of developing Arabidopsis embryos.
Hinz G, Colanesi S, Hillmer S, Rogers JC, Robinson DG. Hinz G, et al. Traffic. 2007 Oct;8(10):1452-64. doi: 10.1111/j.1600-0854.2007.00625.x. Epub 2007 Aug 13. Traffic. 2007. PMID: 17696967 - The rice RMR1 associates with a distinct prevacuolar compartment for the protein storage vacuole pathway.
Shen Y, Wang J, Ding Y, Lo SW, Gouzerh G, Neuhaus JM, Jiang L. Shen Y, et al. Mol Plant. 2011 Sep;4(5):854-68. doi: 10.1093/mp/ssr025. Epub 2011 Apr 14. Mol Plant. 2011. PMID: 21493745 - Pathways for protein transport to seed storage vacuoles.
Jolliffe NA, Craddock CP, Frigerio L. Jolliffe NA, et al. Biochem Soc Trans. 2005 Nov;33(Pt 5):1016-8. doi: 10.1042/BST20051016. Biochem Soc Trans. 2005. PMID: 16246035 Review. - Biogenesis of Plant Prevacuolar Multivesicular Bodies.
Cui Y, Shen J, Gao C, Zhuang X, Wang J, Jiang L. Cui Y, et al. Mol Plant. 2016 Jun 6;9(6):774-86. doi: 10.1016/j.molp.2016.01.011. Epub 2016 Feb 2. Mol Plant. 2016. PMID: 26836198 Review.
Cited by
- Fluorescent reporter proteins for the tonoplast and the vacuolar lumen identify a single vacuolar compartment in Arabidopsis cells.
Hunter PR, Craddock CP, Di Benedetto S, Roberts LM, Frigerio L. Hunter PR, et al. Plant Physiol. 2007 Dec;145(4):1371-82. doi: 10.1104/pp.107.103945. Epub 2007 Sep 28. Plant Physiol. 2007. PMID: 17905861 Free PMC article. - Structural insights into how vacuolar sorting receptors recognize the sorting determinants of seed storage proteins.
Tsao HE, Lui SN, Lo AH, Chen S, Wong HY, Wong CK, Jiang L, Wong KB. Tsao HE, et al. Proc Natl Acad Sci U S A. 2022 Jan 4;119(1):e2111281119. doi: 10.1073/pnas.2111281119. Proc Natl Acad Sci U S A. 2022. PMID: 34983843 Free PMC article. - The formation, function and fate of protein storage compartments in seeds.
Ibl V, Stoger E. Ibl V, et al. Protoplasma. 2012 Apr;249(2):379-92. doi: 10.1007/s00709-011-0288-z. Epub 2011 May 26. Protoplasma. 2012. PMID: 21614590 - GLUTELIN PRECURSOR ACCUMULATION3 encodes a regulator of post-Golgi vesicular traffic essential for vacuolar protein sorting in rice endosperm.
Ren Y, Wang Y, Liu F, Zhou K, Ding Y, Zhou F, Wang Y, Liu K, Gan L, Ma W, Han X, Zhang X, Guo X, Wu F, Cheng Z, Wang J, Lei C, Lin Q, Jiang L, Wu C, Bao Y, Wang H, Wan J. Ren Y, et al. Plant Cell. 2014 Jan;26(1):410-25. doi: 10.1105/tpc.113.121376. Epub 2014 Jan 31. Plant Cell. 2014. PMID: 24488962 Free PMC article. - SCAMPs highlight the developing cell plate during cytokinesis in tobacco BY-2 cells.
Lam SK, Cai Y, Hillmer S, Robinson DG, Jiang L. Lam SK, et al. Plant Physiol. 2008 Aug;147(4):1637-45. doi: 10.1104/pp.108.119925. Epub 2008 May 28. Plant Physiol. 2008. PMID: 18508957 Free PMC article.
References
- Castelli, S., and Vitale, A. (2005). The phaseolin vacuolar sorting signal promotes transient, strong membrane association and aggregation of the bean storage protein in transgenic tobacco. J. Exp. Bot. 56 1379–1387. - PubMed
- Chen, X., Pfeil, J.E., and Gal, S. (2002). The three aspartic proteinase genes of Arabidopsis thaliana are differentially expressed. Eur. J. Biochem. 269 4675–4684. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials