Glycan microarray technologies: tools to survey host specificity of influenza viruses - PubMed (original) (raw)
Review
Glycan microarray technologies: tools to survey host specificity of influenza viruses
James Stevens et al. Nat Rev Microbiol. 2006 Nov.
Abstract
New technologies are urgently required for rapid surveillance of the current H5N1 avian influenza A outbreaks to gauge the potential for adaptation of the virus to the human population, a crucial step in the emergence of pandemic influenza virus strains. Owing to the species-specific nature of the interaction between the virus and host glycans, attention has recently focused on novel glycan array technologies that can rapidly assess virus receptor specificity and the potential emergence of human-adapted H5N1 viruses.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
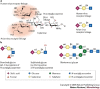
Figure 1. Glycans and influenza virus specificity.
a | The two possible positions of the sialic-acid linkage to a vicinal galactose (α2-6 and α2-3), which are crucial for recognition by the haemagglutinin (HA) proteins of avian and human viruses, are shown in the left panel. The sialic-acid moiety is highlighted by red shading. Cartoon representations of the typical sialic-acid linkage that is bound by avian and human influenza virus HAs are shown in the right panel. b | Cartoon representations of some of the different glycans used in the microarray experiments and discussed in the main text are shown. To simplify glycan descriptions, a symbol and text nomenclature has been developed by the
Nomenclature Committee of the Consortium for Functional Glycomics
.
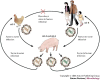
Figure 2. Known mechanisms for the emergence of pandemic influenza A virus strains.
Two of the three pandemic influenza A virus strains during the past century, H2N2 in 1957 (which caused Asian flu) and H3N2 in 1968 (which caused Hong Kong flu), arose from genetic reassortment, in which gene segments from an avian virus were mixed with genetic material from a co-infecting human virus, probably through an intermediate host, such as a pig (the mixing vessel theory). The haemagglutinin (HA) of human influenza A viruses has a binding preference for cell receptors that contain α2-6-linked sialic-acid moieties, whereas avian viruses bind preferentially to α2-3-linked sialic acids moieties. The respiratory epithelial cells of pigs have receptors that express both α2-3- and α2-6-linked sialic-acid moieties, and can be infected by both human and avian viruses. The resulting viral progeny will either be intact avian or human virus, which can only infect their respective hosts or, if reassortment yields functional virus (usually swapped HA, PB2 and/or neuraminidase (NA)), a new pandemic strain might emerge with the ability to retain efficient human-to-human transfer, but be sufficiently different (for example, by a species change in HA) to reduce the ability of the host to mount an effective immune response. PB2, polymerase basic-2 protein.
Figure 3. Schematic representation of the two assays for analysis of the influenza A virus receptor-binding domain.
Recombinant haemagglutinin (HA) analysis involves cloning, expression and purification of recombinant viral HA, with the incorporation of a histidine (His) tag. Approximately 200 different glycans are bound to a glass slide, and the binding of recombinant HA proteins to the different glycans is detected by fluorescent antibodies that bind to a His tag on the recombinant HA. Preliminary experiments used His-tagged recombinant HA pre-complexed to a mouse fluorescent anti-penta-His antibody (six HA-binding sites per complex), but this failed to detect binding to the array owing to the weak HA affinity (mM range) for receptor analogues. Binding to a secondary fluorescent anti-mouse-IgG1 antibody (resulting in twelve HA-binding sites per complex) allowed the detection of fluorescence. It was concluded that the additional multivalency enabled detection through avidity effects, comparable to the natural interaction of HA trimers on the viral envelope with the host cell. The intensity of the fluorescent signal determines whether the binding is recorded as strong, weak or absent. The HA produced by this method can also be used for crystallization and structural studies. Whole-virus analysis involves glycan microarray analysis with intact virus. IEX, ion-exchange chromatography; NA, neuraminidase; M2, M2 ion-channel protein; SEC, size exclusion chromatography.
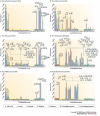
Figure 4. Glycan microarray analysis of human H1, human H3, avian H5 and duck H3 influenza virus haemagglutinins.
Glycan binding analyses as measured by fluorescence intensity are presented for natural haemagglutinins (HAs) from influenza viruses circulating during the 1918 pandemic (A/South Carolina/1/1918 (a) and A/New York/1/1918 (b)), a human H1 (A/Texas/1991) (c), an avian H5 originally isolated from a 10-year-old Vietnamese boy who died from bird flu (A/Vietnam/1203/2004) (d), a human H3 (A/Moscow/10/1999) (e) and a duck H3 (A/Duck/Ukraine/1963), the probable progenitor of the pandemic 1968 Hong Kong virus (f). Binding results for glycoproteins (highlighted in red), α2-3-linked sialosides (highlighted in yellow) and α2-6-linked sialosides (highlighted in green) are shown. The bars corresponding to signals generated by binding to sulphated or additional negatively charged glycans are coloured red. Owing to continual glycan microarray development, a number of new ligands were printed for the analysis of A/Vietnam/1203/2004 HA. Therefore, binding to glycans 37–44 and 58–60 was not determined, except for the A/Vietnam/1203/2004. Cartoon representations of the glycans bound to different spots on the array (numbered 1 to 60) are shown. AGP, α1-acid glycoprotein. Data adapted from REF. and REF. .
Similar articles
- Preferential recognition of avian-like receptors in human influenza A H7N9 viruses.
Xu R, de Vries RP, Zhu X, Nycholat CM, McBride R, Yu W, Paulson JC, Wilson IA. Xu R, et al. Science. 2013 Dec 6;342(6163):1230-5. doi: 10.1126/science.1243761. Science. 2013. PMID: 24311689 Free PMC article. - Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus.
Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. Stevens J, et al. Science. 2006 Apr 21;312(5772):404-10. doi: 10.1126/science.1124513. Epub 2006 Mar 16. Science. 2006. PMID: 16543414 - H5N1 receptor specificity as a factor in pandemic risk.
Paulson JC, de Vries RP. Paulson JC, et al. Virus Res. 2013 Dec 5;178(1):99-113. doi: 10.1016/j.virusres.2013.02.015. Epub 2013 Apr 22. Virus Res. 2013. PMID: 23619279 Free PMC article. Review. - Adaptation of influenza viruses to human airway receptors.
Thompson AJ, Paulson JC. Thompson AJ, et al. J Biol Chem. 2021 Jan-Jun;296:100017. doi: 10.1074/jbc.REV120.013309. Epub 2020 Nov 22. J Biol Chem. 2021. PMID: 33144323 Free PMC article. Review. - Enhanced Human-Type Receptor Binding by Ferret-Transmissible H5N1 with a K193T Mutation.
Peng W, Bouwman KM, McBride R, Grant OC, Woods RJ, Verheije MH, Paulson JC, de Vries RP. Peng W, et al. J Virol. 2018 Apr 27;92(10):e02016-17. doi: 10.1128/JVI.02016-17. Print 2018 May 15. J Virol. 2018. PMID: 29491160 Free PMC article.
Cited by
- Oxidative Release of Natural Glycans: Unraveling the Mechanism for Rapid N-Glycan Glycomics Analysis.
Zhang Q, Lasanajak Y, Song X. Zhang Q, et al. Anal Chem. 2024 Oct 22;96(42):16750-16757. doi: 10.1021/acs.analchem.4c03246. Epub 2024 Oct 10. Anal Chem. 2024. PMID: 39387489 Free PMC article. - Unraveling dynamics of paramyxovirus-receptor interactions using nanoparticles displaying hemagglutinin-neuraminidase.
Wu X, Goebbels M, Debski-Antoniak O, Marougka K, Chao L, Smits T, Wennekes T, Kuppeveld FJMV, Vries E, de Haan CAM. Wu X, et al. PLoS Pathog. 2024 Jul 25;20(7):e1012371. doi: 10.1371/journal.ppat.1012371. eCollection 2024 Jul. PLoS Pathog. 2024. PMID: 39052678 Free PMC article. - Benzene with Alkyl Chains Is a Universal Scaffold for Multivalent Virucidal Antivirals.
Zhu Y, Gasbarri M, Zebret S, Pawar S, Mathez G, Diderich J, Valencia-Camargo AD, Russenberger D, Wang H, Silva PHJ, Dela Cruz JB, Wei L, Cagno V, Münz C, Speck RF, Desmecht D, Stellacci F. Zhu Y, et al. ACS Cent Sci. 2024 Apr 4;10(5):1012-1021. doi: 10.1021/acscentsci.4c00054. eCollection 2024 May 22. ACS Cent Sci. 2024. PMID: 38799657 Free PMC article. - Asymmetrical Biantennary Glycans Prepared by a Stop-and-Go Strategy Reveal Receptor Binding Evolution of Human Influenza A Viruses.
Ma S, Liu L, Eggink D, Herfst S, Fouchier RAM, de Vries RP, Boons GJ. Ma S, et al. JACS Au. 2024 Jan 23;4(2):607-618. doi: 10.1021/jacsau.3c00695. eCollection 2024 Feb 26. JACS Au. 2024. PMID: 38425896 Free PMC article. - Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Thompson AJ, Wu NC, Canales A, Kikuchi C, Zhu X, de Toro BF, Cañada FJ, Worth C, Wang S, McBride R, Peng W, Nycholat CM, Jiménez-Barbero J, Wilson IA, Paulson JC. Thompson AJ, et al. Cell Host Microbe. 2024 Feb 14;32(2):261-275.e4. doi: 10.1016/j.chom.2024.01.003. Epub 2024 Feb 1. Cell Host Microbe. 2024. PMID: 38307019 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical