Phylogenomics of the genus Mus (Rodentia; Muridae): extensive genome repatterning is not restricted to the house mouse - PubMed (original) (raw)
Phylogenomics of the genus Mus (Rodentia; Muridae): extensive genome repatterning is not restricted to the house mouse
Frederic Veyrunes et al. Proc Biol Sci. 2006.
Abstract
The house mouse (Mus musculus) is universally adopted as the mammalian laboratory model, and it is involved in most studies of large-scale comparative genomics. Paradoxically, this taxon is rarely the index species for evolutionary analyses of genome architecture owing to its highly rearranged karyotype. To unravel the origin and nature of this extensive repatterning genome, we performed a multidirectional chromosome painting study of representative species within the genus Mus. However, the latter includes four extant subgenera (Mus, Coelomys, Nannomys and Pyromys) between which the phylogenetic relationships remain elusive despite the numerous molecular studies. Comparative genomic maps were established using chromosome-specific painting probes of the laboratory mouse and Nannomys minutoides. Hence, by integrating closely related species within Mus, this study allowed us to: (i) unambiguously resolve for the first time the long-standing controversial phylogeny, (ii) trace the evolution of genome organization in the house mouse, (iii) track rearrangements that necessitated new centromere locations, i.e. formation of neocentromere or reactivation of latent centromeres, (iv) reveal an extremely high rate of karyotypic evolution, with a 10- to 30-fold acceleration which was coincidental with subgeneric cladogenesis and (v) highlight genomic areas of interest for high-resolution studies on neocentromere formation and synteny breakpoints.
Figures
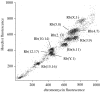
Figure 1
Flow karyotype of a male Nannomys minutoides (2_n_=18) resolving 10 peaks, each containing an Rb fusion chromosome pair.
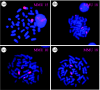
Figure 2
Examples of hybridization using mouse chromosome paints (MMU) to (a,b) Nannomys mattheyi and (c,d) Coelomys pahari metaphase spreads counterstained with DAPI.
Figure 3
G-banded karyotypes of (a) Nannomys mattheyi and (b) Coelomys pahari with the assignments of Mus musculus homologous segments (chromosome pair numbers on the right) revealed by the mouse probes. ‘dist’, ‘med’ and ‘prox’ refer to the distal, median and proximal segment of the chromosome, respectively, and a and b refer to unidentified subchromosomal segments.
Figure 4
Most parsimonious phylogeny using PAUP, based on the 67 chromosomal characters (electronic supplementary material). Bootstrap values supporting each clade are indicated in bold on nodes. The chromosomal rearrangements, which have occurred within the genus Mus are mapped onto the tree and are numbered in grey. Each rearrangement is coded as follows: transl, translocation; inv, inversion; fiss, fission; fus, fusion followed by the character numbers of the table in the electronic supplementary material. Asterisk indicates the emergence of a new centromere. Rearrangements 1–3: transl* + inv 52–53; fiss* 3—rearrangements 4–11: inv 14; fus 16; fus 8; fus 58; transl 11; transl* 60; transl* 10; transl* 2—rearrangements 12–14: transl* 57; transl* 56; fus 59—rearrangements 15–19: transl* 1; transl* 18; transl 5; transl* 4; fus 13—rearrangements 20–22: fiss* 25; fiss* 26; fiss* 58—rearrangements 23–29: transl* 66; transl* 65; transl* 67; fus 64; fus 62; fus 61; fus 63. The divergence times mentioned follow Chevret et al. (2005) and Veyrunes et al. (2005).
Figure 5
Inferred haploid set of the ancestral karyotype of the genus Mus (2_n_=46) reconstructed with Coelomys, Nannomys and Mus G-banded chromosomes. Homology to Mus chromosomes is indicated to the right of each putative ancestral chromosome. ‘dist’, ‘med’ and ‘prox’ refer to the distal, median and proximal segment of the chromosome, respectively, and a and b refer to unidentified subchromosomal segments.
Similar articles
- Are ribosomal DNA clusters rearrangement hotspots?: a case study in the genus Mus (Rodentia, Muridae).
Cazaux B, Catalan J, Veyrunes F, Douzery EJ, Britton-Davidian J. Cazaux B, et al. BMC Evol Biol. 2011 May 13;11:124. doi: 10.1186/1471-2148-11-124. BMC Evol Biol. 2011. PMID: 21569527 Free PMC article. - Phylogenetic relationships in the genus mus, based on paternally, maternally, and biparentally inherited characters.
Lundrigan BL, Jansa SA, Tucker PK. Lundrigan BL, et al. Syst Biol. 2002 Jun;51(3):410-31. doi: 10.1080/10635150290069878. Syst Biol. 2002. PMID: 12079642 - Praomys tullbergi (Muridae, Rodentia) genome architecture decoded by comparative chromosome painting with Mus and Rattus.
Chaves R, Louzada S, Meles S, Wienberg J, Adega F. Chaves R, et al. Chromosome Res. 2012 Aug;20(6):673-83. doi: 10.1007/s10577-012-9304-1. Epub 2012 Jul 31. Chromosome Res. 2012. PMID: 22847644 - Influence of Cat Odor on Reproductive Behavior and Physiology in the House Mouse: (Mus Musculus).
Voznessenskaya VV. Voznessenskaya VV. In: Mucignat-Caretta C, editor. Neurobiology of Chemical Communication. Boca Raton (FL): CRC Press/Taylor & Francis; 2014. Chapter 14. In: Mucignat-Caretta C, editor. Neurobiology of Chemical Communication. Boca Raton (FL): CRC Press/Taylor & Francis; 2014. Chapter 14. PMID: 24830030 Free Books & Documents. Review. - Evolutionary molecular cytogenetics of catarrhine primates: past, present and future.
Stanyon R, Rocchi M, Bigoni F, Archidiacono N. Stanyon R, et al. Cytogenet Genome Res. 2012;137(2-4):273-84. doi: 10.1159/000339381. Epub 2012 Jun 16. Cytogenet Genome Res. 2012. PMID: 22710640 Review.
Cited by
- IFI207, a young and fast-evolving protein, controls retroviral replication via the STING pathway.
Moran EA, Salas-Briceno K, Zhao W, Enya T, Aguilera AN, Acosta I, Alonzo F, Kiani D, Behnsen J, Alvarez C, Keane TM, Adams DJ, Lilue J, Ross SR. Moran EA, et al. mBio. 2024 Jul 17;15(7):e0120924. doi: 10.1128/mbio.01209-24. Epub 2024 Jun 11. mBio. 2024. PMID: 38860764 Free PMC article. - Two genetic determinants acquired late in Mus evolution regulate the inclusion of exon 5, which alters mouse APOBEC3 translation efficiency.
Li J, Hakata Y, Takeda E, Liu Q, Iwatani Y, Kozak CA, Miyazawa M. Li J, et al. PLoS Pathog. 2012 Jan;8(1):e1002478. doi: 10.1371/journal.ppat.1002478. Epub 2012 Jan 19. PLoS Pathog. 2012. PMID: 22275865 Free PMC article. - Adaptive evolution of Mus Apobec3 includes retroviral insertion and positive selection at two clusters of residues flanking the substrate groove.
Sanville B, Dolan MA, Wollenberg K, Yan Y, Martin C, Yeung ML, Strebel K, Buckler-White A, Kozak CA. Sanville B, et al. PLoS Pathog. 2010 Jul 1;6(7):e1000974. doi: 10.1371/journal.ppat.1000974. PLoS Pathog. 2010. PMID: 20617165 Free PMC article. - The mouse "xenotropic" gammaretroviruses and their XPR1 receptor.
Kozak CA. Kozak CA. Retrovirology. 2010 Nov 30;7:101. doi: 10.1186/1742-4690-7-101. Retrovirology. 2010. PMID: 21118532 Free PMC article. Review. - Proteomic study of the brackish water mussel Mytilopsis leucophaeata.
Stekhoven FMS, van der Velde G, Lee TH, Bottrill AR. Stekhoven FMS, et al. Zool Stud. 2015 Jan 23;54:e22. doi: 10.1186/s40555-014-0081-8. eCollection 2015. Zool Stud. 2015. PMID: 31966109 Free PMC article.
References
- Amor D.J, Bentley K, Ryan J, Perry J, Wong L, Slater H, Choo K.H.A. Human centromere repositioning “in progress”. Proc. Natl Acad. Sci. USA. 2004;101:6542–6547. doi:10.1073/pnas.0308637101 - DOI - PMC - PubMed
- Bailey J.A, Baertsch R, James Kent W, Haussler D, Eichler E.E. Hotspots of mammalian chromosomal evolution. Genome Biol. 2004;5:R23. doi:10.1186/gb-2004-5-4-r23 - DOI - PMC - PubMed
- Bonhomme F. Evolutionary relationships in the genus Mus. Curr. Top. Microbiol. Immun. 1986;127:19–34. - PubMed
- Bonhomme F. Genetic diversity and evolution in the genus Mus. In: Goldowitz W.D, Winner R.E, editors. Techniques for the genetic analysis of brain and behavior. Elsevier Sciences Publishers, BV; North Holland, NY: 1992.
- Boursot P, Auffray J.C, Britton-Davidian J, Bonhomme F. The evolution of house mice. Annu. Rev. Ecol. Syst. 1993;24:119–152. doi:10.1146/annurev.es.24.110193.001003 - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources