Cracking the egg: molecular dynamics and evolutionary aspects of the transition from the fully grown oocyte to embryo - PubMed (original) (raw)
Cracking the egg: molecular dynamics and evolutionary aspects of the transition from the fully grown oocyte to embryo
Alexei V Evsikov et al. Genes Dev. 2006.
Abstract
Fully grown oocytes (FGOs) contain all the necessary transcripts to activate molecular pathways underlying the oocyte-to-embryo transition (OET). To elucidate this critical period of development, an extensive survey of the FGO transcriptome was performed by analyzing 19,000 expressed sequence tags of the Mus musculus FGO cDNA library. Expression of 5400 genes and transposable elements is reported. For a majority of genes expressed in mouse FGOs, homologs transcribed in eggs of Xenopus laevis or Ciona intestinalis were found, pinpointing evolutionary conservation of most regulatory cascades underlying the OET in chordates. A large proportion of identified genes belongs to several gene families with oocyte-restricted expression, a likely result of lineage-specific genomic duplications. Gene loss by mutation and expression in female germline of retrotransposed genes specific to M. musculus is documented. These findings indicate rapid diversification of genes involved in female reproduction. Comparison of the FGO and two-cell embryo transcriptomes demarcated the processes important for oogenesis from those involved in OET and identified novel motifs in maternal mRNAs associated with transcript stability. Discovery of oocyte-specific eukaryotic translation initiation factor 4E distinguishes a novel system of translational regulation. These results implicate conserved pathways underlying transition from oogenesis to initiation of development and illustrate how genes acquire and lose reproductive functions during evolution, a potential mechanism for reproductive isolation.
Figures
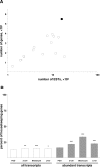
FIGURE 1.
Complexity of the FGO cDNA library. (A) Numbers of genes represented by ESTs of the FGO library (black circle) and 15 cDNA libraries of oocytes and preimplantation embryos (white circles). (_X_-axis) Number of ESTs corresponding to genes in the libraries; (_Y_-axis) number of genes these ESTs represent. (B) Fraction of housekeeping genes among all (left) and abundant (right) transcripts identified in the FGOs, two-cell stage embryos, blastocysts, and liver. (*) p < 0.05; (**) p < 0.01; (***) p <0.001.
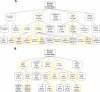
FIGURE 2.
Structure of the FGO transcriptome. Biological process (A) and molecular function (B) GO annotations common to at least 5% of FGO genes. The top three levels of GO hierarchy and the numbers of FGO genes annotated with a corresponding GO term are shown. GO terms associated with housekeeping genes are in boxes with yellow borders. Dotted connector lines indicate that a GO term is a subset of two distinct higher-level terms.
FIGURE 3.
Analysis of transcripts identified in the FGO library. (A) RT–PCR detection of Nalp family (Nalp5, Nalp2, Nalp14, Nalp4f), Eif4eloo, sense and antisense Suv39h1 (Suv39h1, Suv39h1as), and Zp4 transcripts. (FGO) Fully grown oocyte; (OO) ovulated oocyte; (Zyg) zygote; (e2C) early two-cell stage embryo; (l2C) late two-cell stage embryo; (8c) early eight-cell stage embryo; (Mor) morula; (Bl) blastocyst; (H2O) control for PCR reactions; (−RT) reverse transcriptase omitted (FGO sample); (MW) DNA size markers. (B) Alignments of LOC278215 protein with BMI1 of H. sapiens, R. norvegicus, and M. musculus. Identical amino acids are red, and residues conserved in at least half of proteins are blue. (C) Partial alignment of M. musculus, R. norvegicus, and H. sapiens Zp4 cDNA sequences. Second and fourth exons are in lowercase. Start codons are underlined in green, and stop codons in the M. musculus Zp4 are underlined in red.
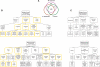
FIGURE 4.
Conservation of gene expression in the oocytes of Chordata. (A) Homologs of genes expressed in M. musculus FGOs are transcribed in eggs of X. laevis and C. intestinalis. (B) Genes with conserved expression are biased to specific biological processes and molecular functions. (*) p < 0.01; (**) p < 0.001; (***) p < 0.0001. Boxes with yellow borders and dotted connector lines are as in Figure 2A. Higher-level GO terms for overrepresented categories (boxes with dotted borders) are shown for reference. (C) Biological processes and molecular functions overrepresented among _M. musculus_-specific FGO-expressed genes. Presentation of data is as in B.
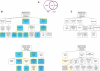
FIGURE 5.
Gene expression during the OET. (A) Comparison of the FGO and two-cell stage mouse embryo transcriptomes. (B) GO categories overrepresented among genes identified in both cDNA libraries. Presentation of data is as in Figure 4B. Blue boxes indicate that the same GO terms are overrepresented among genes whose expression is conserved in the eggs of chordates. (C) GO categories overrepresented among FGO genes absent in the two-cell stage embryo. Presentation of data is as in Figure 4B.
FIGURE 6.
Translational regulation of maternal mRNAs. (A) Cumulative distribution of 3′-UTR lengths for transient, stable, and activating transcripts. (B) Sequence logo representation of motif consensi identified by reciprocal analysis in the transient (motifs 1–4) and stable (motifs 5–8) mRNAs. (C) Exon–intron structure for Eif4eloo transcript variants identified by their GenBank accession numbers (all transcripts but DQ279473 are incomplete at the 3′ end). (D) Alignment for a major isoform of the M. musculus EIF4ELOO with predicted proteins from Monodelphis domestica, H. sapiens, Macaca mulatta, Bos taurus, Canis familiaris, and R. norvegicus. Color coding is as in Figure 3B. Gray shading is the IF4E/CDC33 domain.
Similar articles
- Stable maternal proteins underlie distinct transcriptome, translatome, and proteome reprogramming during mouse oocyte-to-embryo transition.
Zhang H, Ji S, Zhang K, Chen Y, Ming J, Kong F, Wang L, Wang S, Zou Z, Xiong Z, Xu K, Lin Z, Huang B, Liu L, Fan Q, Jin S, Deng H, Xie W. Zhang H, et al. Genome Biol. 2023 Jul 13;24(1):166. doi: 10.1186/s13059-023-02997-8. Genome Biol. 2023. PMID: 37443062 Free PMC article. - A novel maternally transcribed homeobox gene, Eso-1, is preferentially expressed in oocytes and regulated by cytoplasmic polyadenylation.
Li H, Tsai MS, Chen CY, Lian WC, Chiu YT, Chen GD, Wang SH. Li H, et al. Mol Reprod Dev. 2006 Jul;73(7):825-33. doi: 10.1002/mrd.20478. Mol Reprod Dev. 2006. PMID: 16596637 - Controlling the Messenger: Regulated Translation of Maternal mRNAs in Xenopus laevis Development.
Sheets MD, Fox CA, Dowdle ME, Blaser SI, Chung A, Park S. Sheets MD, et al. Adv Exp Med Biol. 2017;953:49-82. doi: 10.1007/978-3-319-46095-6_2. Adv Exp Med Biol. 2017. PMID: 27975270 Free PMC article. Review. - Translational Regulation in the Mammalian Oocyte.
Susor A, Kubelka M. Susor A, et al. Results Probl Cell Differ. 2017;63:257-295. doi: 10.1007/978-3-319-60855-6_12. Results Probl Cell Differ. 2017. PMID: 28779322 Review.
Cited by
- miR-135A regulates preimplantation embryo development through down-regulation of E3 Ubiquitin Ligase Seven In Absentia Homolog 1A (SIAH1A) expression.
Pang RT, Liu WM, Leung CO, Ye TM, Kwan PC, Lee KF, Yeung WS. Pang RT, et al. PLoS One. 2011;6(11):e27878. doi: 10.1371/journal.pone.0027878. Epub 2011 Nov 22. PLoS One. 2011. PMID: 22132158 Free PMC article. - The oocyte-to-embryo transition in mouse: past, present, and future.
Schultz RM, Stein P, Svoboda P. Schultz RM, et al. Biol Reprod. 2018 Jul 1;99(1):160-174. doi: 10.1093/biolre/ioy013. Biol Reprod. 2018. PMID: 29462259 Free PMC article. - Wispy, the Drosophila homolog of GLD-2, is required during oogenesis and egg activation.
Cui J, Sackton KL, Horner VL, Kumar KE, Wolfner MF. Cui J, et al. Genetics. 2008 Apr;178(4):2017-29. doi: 10.1534/genetics.107.084558. Genetics. 2008. PMID: 18430932 Free PMC article. - Transcriptome Analysis of ESTs from a Chaetognath Reveals a Deep-Branching Clade of Retrovirus-Like Retrotransposons.
Barthélémy RM, Casanova JP, Faure E. Barthélémy RM, et al. Open Virol J. 2008;2:44-60. doi: 10.2174/1874357900802010044. Epub 2008 May 7. Open Virol J. 2008. PMID: 19440464 Free PMC article. - Comparative Proteomic Profiling Reveals Molecular Characteristics Associated with Oogenesis and Oocyte Maturation during Ovarian Development of Bactrocera dorsalis (Hendel).
Wei D, Li R, Zhang MY, Liu YW, Zhang Z, Smagghe G, Wang JJ. Wei D, et al. Int J Mol Sci. 2017 Jun 30;18(7):1379. doi: 10.3390/ijms18071379. Int J Mol Sci. 2017. PMID: 28665301 Free PMC article.
References
- Barreau, C., Paillard, L., Mereau, A., Osborne, H.B. Mammalian CELF/Bruno-like RNA-binding proteins: Molecular characteristics and biological functions. Biochimie. 2006;88:515–525. - PubMed
- Bultman, S., Magnuson, T. Molecular and genetic analysis of the mouse homolog of the Drosophila suppressor of position-effect variegation 3–9 gene. Mamm. Genome. 2000;11:251–254. - PubMed
- Byrne, J.A., Simonsson, S., Western, P.S., Gurdon, J.B. Nuclei of adult mammalian somatic cells are directly reprogrammed to oct-4 stem cell gene expression by amphibian oocytes. Curr. Biol. 2003;13:1206–1213. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HD037102/HD/NICHD NIH HHS/United States
- P30 CA034196/CA/NCI NIH HHS/United States
- P20 RR016463/RR/NCRR NIH HHS/United States
- P20 RR16463/RR/NCRR NIH HHS/United States
- HD021970/HD/NICHD NIH HHS/United States
- CA34196/CA/NCI NIH HHS/United States
- HD037102/HD/NICHD NIH HHS/United States
- R37 HD021970/HD/NICHD NIH HHS/United States
- U01 HD021970/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases