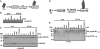ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome - PubMed (original) (raw)
ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome
Chang-Wei Liu et al. Mol Cell. 2006.
Abstract
The 26S proteasome degrades polyubiquitinated proteins by an energy-dependent mechanism. Here we define multiple roles for ATP in 26S proteasome function. ATP binding is necessary and sufficient for assembly of 26S proteasome from 20S proteasome and PA700/19S subcomplexes and for proteasome activation. Proteasome assembly and activation may require distinct ATP binding events. The 26S proteasome degrades nonubiquitylated, unstructured proteins without ATP hydrolysis, indicating that substrate translocation per se does not require the energy of hydrolysis. Nonubiquitylated folded proteins and certain polyubiquitylated folded proteins were refractory to proteolysis. The latter were deubiquitylated by an ATP-independent mechanism. Other folded as well as unstructured polyubiquitylated proteins required ATP hydrolysis for proteolysis and deubiquitylation. Thus, ATP hydrolysis is not used solely for substrate unfolding. These results indicate that 26S proteasome-catalyzed degradation of polyubiquitylated proteins involves mechanistic coupling of several processes and that such coupling imposes an energy requirement not apparent for any isolated process.
Figures
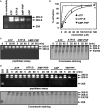
Figure 1. Characterization of Purified 26S Proteasome from Bovine Blood Red Cells
(A) 26S proteasome (960 ng), PA700 (400 ng), and 20S proteasome (260 ng) were subjected to SDS-PAGE and visualized by silver staining. (B) 26S proteasome (3 mg), PA700 (3 mg), and 20S proteasome (3 mg) were subjected to 4% native PAGE and either stained with Coomassie blue (upper) or assayed for proteasome activity with an overlay assay (lower) using Suc-Leu-Leu-Val-Tyr-AMC as a fluorogenic substrate. (C) Assembly of doubly capped 26S proteasome (26S-D) from singly capped 26S proteasome (26S-S) and PA700. Predominately singly capped 26S proteasome (40 nM) was incubated with purified PA700 (400 nM) in the presence of 200 μM ATP for the indicated times and subjected to 4% native PAGE. Gels were either stained with Coomassie blue (upper) or assayed for proteasome activity with a substrate overlay assay (lower). Migration positions of purified 20S and PA700 were established as references by electrophoresis of respective proteins.
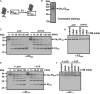
Figure 2. ATP Binding Is Necessary and Sufficient for Assembly of 26S Proteasome from PA700 and 20S Proteasome and for 26S Proteasome Stability
(A) ATP and nonhydrolyzable ATP analogs promote assembly and activation of the 26S proteasome. 20S proteasome (40 nM) was incubated with PA700 (320 nM) for 60 min with no nucleotide or in the presence of 1 mM ATP, ATPγS, AMP-PNP, or ADP. Samples were subjected to a native gel electrophoresis and assayed for proteasome activity with a substrate overlay assay by using Suc-Leu-Leu-Val-Tyr-AMC. (B) Assembly reactions were as described in (A) at the indicated nucleotide concentrations. Soluble proteasome assays were conducted with Suc-Leu-Leu-Val-Tyr-AMC. Results are expressed as a percentage of maximal activation for each nucleotide. ATP, ATPγS, and AMP-PNP maximally activated proteasome activity by 19.8-, 10.1-, and 4.4-fold, respectively. No activation of 20S proteasome by PA700 was achieved in the absence of nucleotides. (C) Nucleotide binding-dependent assembly of doubly capped 26S from singly capped 26S proteasome and PA700. Purified 26S proteasome (40 nM, approximately 60% doubly capped versus 40% singly capped) was incubated with PA700 (120 nM) in buffer A at 37°C for the indicated times. In reactions with ATPγS, AMP-PNP, or no ATP (2ATP), 26S proteasome was preincubated for 1 min at 30°C with apyrase (8 mU/μl) prior to the addition of ATPγS, AMP-PNP (1 mM), or buffer, respectively. Samples were subjected to native gel electrophoresis and assayed for proteasome activity using Suc-Leu-Leu-Val-Tyr-AMC (left panel) or stained with Coomassie blue (right panel). (D) 26S proteasome (40 nM) was incubated at 37°C in the presence of 1 mM ATP or treated with 8 mU/μl apyrase for 1 min at 30°C prior to supplementation with 1 mM ATPγS, 1 mM AMP-PNP, 1 mM ADP, or buffer and incubated at 37°C. At the indicated times, samples were subjected to native PAGE and either assayed for proteasome activity by using Suc-Leu-Leu-Val-Tyr-AMC (upper) or stained with Coomassie blue (lower). Purified, nonincubated 20S and 26S proteasomes were electrophoresed and processed for reference.
Figure 3. 26S Proteasome Assembly and Activation Require Distinct ATP Binding Events
(A) Depletion of ATP promotes dissociation of 26S proteasome and loss of activity. Purified 26S proteasome (40 nM) was incubated with 8 mU/μl apyrase at 37°C. At the indicated times, samples were subjected to native PAGE and assayed for proteasome activity using a gel overlay assay with Suc-Leu-Leu-Val-Tyr-AMC (upper panel), stained with Coomassie blue (middle panel), or assayed for proteasome activity using a soluble quantitative fluorescence assay with Suc-Leu-Leu-Val-Tyr-AMC (lower panel). The protein levels in the native gel were quantified by densitometry and plotted with the corresponding proteasome activity (lower panel). (B) ATP binding promotes reactivation of ATP-deficient 26S proteasome. 26S proteasome was incubated for 60 min with apyrase as described above and subjected to Superose 6 gel filtration chromatography to separate the 26S proteasome from the dissociated 20S and PA700 subcomplexes. 26S proteasome isolated from the gel filtration column was subjected to native PAGE (upper panel) and to activity assays in the absence or presence of 0.5 mM ATP or 0.5 mM ATPγS (lower panel). Activity is expressed as arbitrary fluorescent units and represents the mean value of triplicate assays (±SEM). Similar results were obtained in three independent experiments.
Figure 4. ATP Hydrolysis Is Not Required for Degradation of Nonubiquitylated Unstructured Proteins by the 26S Proteasome
(A) Degradation of p21cip1 by doubly capped 26S proteasome. Doubly capped 26S proteasome was prepared in the presence of 1 mM ATP, ATPγS, or AMP-PNP. p21cip1 (1.5 μM) was incubated with doubly capped 26S proteasome (40 nM) at 37°C for the indicated times. Lanes denoted MG indicate that proteasomes were preincubated with 100 μM MG132 prior to addition of substrate and initiation of assay. Samples were either subjected to western blotting against p21cip1 antibody (upper gel) or evaluated for integrity of the 26S proteasome during degradation by Coomassie staining after native PAGE (lower gel). Purified 26S proteasome (containing both doubly capped and singly capped proteasome) and 20S proteasome were also electrophoresed as references for unassembled complexes. p21cip1 was quantified by densitometry after western blotting (lower). Similar results were obtained in three independent experiments. (B) Degradation of [methyl-14C] casein by purified 26S proteasome. Reactions contained 40 nM doubly capped 26S proteasome and 0.2 mg/ml of [methyl-14C] casein in the presence of 1 mM ATP, ATPγS, or AMP-PNP. Proteolysis is expressed as the percentage of casein degraded and represents mean values of triplicate assays (±SEM). Samples before and after the reaction were also subjected to native PAGE to evaluate the integrity of the 26S proteasome (lower gel). (C) Degradation of p21cip1 by 20S proteasome and 26S proteasome. p21cip1 was incubated with indicated concentrations of 20S proteasome or doubly capped 26S proteasome for 8 min at 37°C. Residual p21cip1 was quantified by densitometry after western blotting. Proteolysis is expressed as the percent of initial p21cip1 degraded. Values represent means of triplicate assays (±SEM). (D) 26S proteasome does not require ATP hydrolysis to degrade unstructured proteins. ATP was removed from doubly capped 26S proteasome by apyrase. The sample was subjected to glycerol density gradient centrifugation in ATP-deficient buffer. Fraction 1 (12.5% glycerol), fraction 22 (40% glycerol). Samples from fractions were assayed for degradation of p21cip1 or [methyl-14C] casein (data not shown) as described above in the presence or absence of 0.5 mM ATPγ S. Arrows denote sedimentation positions of purified untreated 20S proteasome and 26S proteasomes in separate control experiments.
Figure 5. Polyubiquitin Chain Engagement and Deubiquitylation Do Not Require ATP Hydrolysis
(A) Scheme for preparation of Ub4-E225K. (B) Ub4-E225K was purified by Superdex 200 chromatography; Ub4-E225K (2 μg) was subjected to 10% SDS-PAGE and visualized by Coomassie staining. (C–F) Deubiquitylation of Ub4-E225K by the 26S proteasome and PA700. Deubiquitylation was monitored by blotting with anti-E225K. (C) Ub4-E225K (120 nM) and purified 26S proteasome (80 nM) were incubated at 37°C for indicated times in the presence of 1 mM ATP or 1 mM ATPγS after depletion of ATP by apyrase. (D) Reactions as in (C) after 180 min of incubation with no 26S proteasome (control) or 26S proteasome after 10 min preincubation with 2.5 mM 1,10-phenanthroline (1, 10 phen) or 2 μM ubiquitin aldehyde (Ubal). (E and F) Deubiquitylation of Ub4-E225K by PA700. Reactions are as in (C) and (D) except with 160 nM PA700 without (–) or with (+) 1 mM ATP.
Figure 6. ATP Hydrolysis Is Required for Degradation of a Polyubiquitylated Protein
(A) Scheme for synthesis of Ub4-Ubch10. (B) Nonubiquitinated Ubch10 is not degraded by the 26S proteasome. Reactions contained Ubch10 (100 nM) and 26S proteasome (30 nM) in the presence of 1 mM ATP or ATPγS. In reactions containing epoxomicin, proteasome was preincubated with 100 μM epoxomicin for 10 min prior to initiation, and reactions were stopped at the last time point. (C) ATP hydrolysis-dependent degradation of Ub4-Ubch10 by the 26S proteasome. Doubly capped 26S proteasome (30 nM) was incubated with Ub4-Ubch10 (100 nM) with 1 mM ATP or 1 mM ATPγS for the indicated times. Ubch10 was detected by western blotting. Lanes denoted Ubal, 1,10-phen, and Epox indicate that proteasome was preincubated with either 2 μM ubiquitin aldehyde, 2.5 mM 1,10-phenanthroline, or 100 μM epoxomicin, respectively, for 10 min prior to initiation of reaction. Ubch10 was monitored by western blotting with an anti-Ubch10 antibody. (D) Scheme for synthesis of Ubn-cyclin B11–102. (E) ATP hydrolysis-dependent degradation of Ubn-cyclin B11–102. Degradation of Ubn-cyclin B11–102 (100 nM) was assessed by western blotting with an anti-myc antibody in reactions analogous to (C). Reactions contained 20 nM 26S proteasome in the presence of 1 mM ATP or ATPγS.
Figure 7. A Model for the Roles of ATP in 26S Proteasome Function
The 26S proteasome is composed of the 20S proteasome and the PA700 regulatory complex. Purified PA700 is an ATPase but deubiquitylates polybubiquitylated proteins by an ATP-independent process (1). The 20S proteasome can degrade certain unstructured proteins that promote gating (2). Assembly of the 26S proteasome from 20S proteasome and PA700 requires ATP binding, but not hydrolysis (3). Activation of the 26S proteasome may require a separate ATP binding event (4). Removal of ATP promotes inactivation (5) and disassembly (6) of the 26S proteasome. The 26S proteasome deubiquitylates, by an ATP-independent mechanism, some polyubiquitylated proteins that are refractory to degradation (1). The 26S proteasome degrades certain unstructured, nonubiquitylated proteins without ATP hydrolysis (2). The 26S proteasome degrades both folded and unstructured polyubiquitylated proteins by a process that requires ATP hydrolysis, and it couples mechanisms required for degradation including substrate unfolding (when needed), deubiquitylation, translocation, and degradation (7 and 8).
Similar articles
- Variably modulated gating of the 26S proteasome by ATP and polyubiquitin.
Li X, Demartino GN. Li X, et al. Biochem J. 2009 Jul 15;421(3):397-404. doi: 10.1042/BJ20090528. Biochem J. 2009. PMID: 19435460 Free PMC article. - ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins.
Smith DM, Kafri G, Cheng Y, Ng D, Walz T, Goldberg AL. Smith DM, et al. Mol Cell. 2005 Dec 9;20(5):687-98. doi: 10.1016/j.molcel.2005.10.019. Mol Cell. 2005. PMID: 16337593 - ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle.
Babbitt SE, Kiss A, Deffenbaugh AE, Chang YH, Bailly E, Erdjument-Bromage H, Tempst P, Buranda T, Sklar LA, Baumler J, Gogol E, Skowyra D. Babbitt SE, et al. Cell. 2005 May 20;121(4):553-565. doi: 10.1016/j.cell.2005.03.028. Cell. 2005. PMID: 15907469 Retracted. - Proteasomes and their associated ATPases: a destructive combination.
Smith DM, Benaroudj N, Goldberg A. Smith DM, et al. J Struct Biol. 2006 Oct;156(1):72-83. doi: 10.1016/j.jsb.2006.04.012. Epub 2006 May 8. J Struct Biol. 2006. PMID: 16919475 Review. - Structure, Dynamics and Function of the 26S Proteasome.
Mao Y. Mao Y. Subcell Biochem. 2021;96:1-151. doi: 10.1007/978-3-030-58971-4_1. Subcell Biochem. 2021. PMID: 33252727 Review.
Cited by
- The Time-Resolved Salt Stress Response of _Dunaliella tertiolecta_-A Comprehensive System Biology Perspective.
Keil L, Mehlmer N, Cavelius P, Garbe D, Haack M, Ritz M, Awad D, Brück T. Keil L, et al. Int J Mol Sci. 2023 Oct 19;24(20):15374. doi: 10.3390/ijms242015374. Int J Mol Sci. 2023. PMID: 37895054 Free PMC article. - Proteasome granule formation is regulated through mitochondrial respiration and kinase signaling.
Waite KA, Roelofs J. Waite KA, et al. J Cell Sci. 2022 Sep 1;135(17):jcs259778. doi: 10.1242/jcs.259778. Epub 2022 Sep 7. J Cell Sci. 2022. PMID: 35975718 Free PMC article. - Highly conserved shifts in ubiquitin-proteasome system (UPS) activity drive mitochondrial remodeling during quiescence.
Yue S, Wang L, DeMartino GN, Zhao F, Liu Y, Sieber MH. Yue S, et al. Nat Commun. 2022 Aug 1;13(1):4462. doi: 10.1038/s41467-022-32206-2. Nat Commun. 2022. PMID: 35915093 Free PMC article. - Early Forms of _α_-Synuclein Pathology Are Associated with Neuronal Complex I Deficiency in the Substantia Nigra of Individuals with Parkinson's Disease.
Flønes IH, Nyland H, Sandnes DA, Alves GW, Tysnes OB, Tzoulis C. Flønes IH, et al. Biomolecules. 2022 May 25;12(6):747. doi: 10.3390/biom12060747. Biomolecules. 2022. PMID: 35740871 Free PMC article. - The ATP-dependent Pathways and Human Diseases.
Suwara J, Radzikowska-Cieciura E, Chworos A, Pawlowska R. Suwara J, et al. Curr Med Chem. 2023;30(11):1232-1255. doi: 10.2174/0929867329666220322104552. Curr Med Chem. 2023. PMID: 35319356
References
- Baumeister W, Walz J, Zühl F, Seemüller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. - PubMed
- Benarouodj N, Zwickl P, Seemüller E, Baumeister W, Goldberg AL. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol. Cell. 2003;11:69–78. - PubMed
- Bochtler M, Ditzel L, Groll M, Hartmann C, Huber R. The proteasome. Annu. Rev. Biophys. Biomol. Struct. 1999;28:295–317. - PubMed
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 1996;65:801–847. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK 46181/DK/NIDDK NIH HHS/United States
- R01 DK046181/DK/NIDDK NIH HHS/United States
- DK 49835/DK/NIDDK NIH HHS/United States
- R01 DK049835/DK/NIDDK NIH HHS/United States
- R56 DK046181/DK/NIDDK NIH HHS/United States
- R37 DK049835/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources