Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression - PubMed (original) (raw)
Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression
Yi-Jun Sheu et al. Mol Cell. 2006.
Abstract
Origins of DNA replication are licensed in G1 by recruiting the minichromosome maintenance (MCM) proteins to form a prereplicative complex (pre-RC). Prior to initiation of DNA synthesis from each origin, a preinitiation complex (pre-IC) containing Cdc45 and other proteins is formed. We report that Cdc7-Dbf4 protein kinase (DDK) promotes assembly of a stable Cdc45-MCM complex exclusively on chromatin in S phase. In this complex, Mcm4 is hyperphosphorylated. Studies in vitro using purified DDK and Mcm4 demonstrate that hyperphosphorylation occurs at the Mcm4 N terminus. However, the DDK substrate specificity is conferred by an adjacent DDK-docking domain (DDD), sufficient for facilitating efficient phosphorylation of artificial phosphoacceptors in cis. Genetic evidence suggests that phosphorylation of Mcm4 by DDK is important for timely S phase progression and for cell viability upon overproduction of Cdc45. We suggest that DDK docks on and phosphorylates MCM proteins at licensed origins to promote proper assembly of pre-IC.
Figures
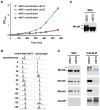
Figure 1. A stable Cdc45-MCM complex containing hyper-phosphorylated Mcm4. Strain OAy535 is used here (Aparicio et al., 1997)
(A) Biochemical fractionation scheme of yeast cells into soluble and chromatin-bound proteins. (B) A Cdc45-MCM complex exists on S-phase chromatin. Left, silver-stained gel of Cdc45 IP (+) or 12CA5 IP (−) of SN2 prepared from cells arrested in G1 (α) and S (HU) phases. Right, results of mass spectrometry for the indicated bands. (C) Cdc45-MCM interaction occurs only on S-phase chromatin. Left, Immuno-blot analysis of SN1 and SN2 from cells arrested in G1 or S phase. Middle, Immuno-blot of Cdc45 IP. Right, Immuno-blot of Mcm4 from Cdc45 IP, SN1 and SN2 fractions. (D) Mcm4 in the Cdc45-MCM complex is hyper-phosphorylated. Immuno-blot analysis of the Mcm4 mobility shift in the Cdc45 IP after phosphatase treatment. (E) Mcm4 hyper-phosphorylation is cell cycle-regulated. Immuno-blot analysis of SN2 samples from the time course of α-factor arrest and release. The percent of unbudded cells is shown. (F) Cdc45-MCM complex formed in S phase contains hyper-phosphorylated Mcm4. Immuno-blot analysis of Cdc45 IP of SN2 from the time-course samples. The small panel at right shows Mcm4 from a Cdc45 IP and SN2 side by side for comparison.
Figure 2. CDC7 is required for normal S-phase progression, hyper-phosphorylation of Mcm4 in vivo and stable Cdc45-MCM complex formation. Strains YS814, YS819, YS824 and YS828 are used here
(A) Growth curve of mcm5-bob1 and mcm5-bob1cdc7Δ cells. (B) S-phase progression of mcm5-bob1 and mcm5-bob1cdc7Δ cells. Cells were arrested in G1 using α-factor, released into fresh YPD medium at room temperature, collected at indicated time points, fixed and analyzed by flow cytometry for DNA content. (C) Hyper-phosphorylation of Mcm4 is not detectable in cdc7Δ cells arrested in HU. Immuno-blot analysis of the Mcm4 mobility shift. 8 μg of protein from SN2 is loaded in each lane. (D) Cdc45-MCM complex is not stable in cdc7Δ cells. Left panel, SN2 from HU arrested cells. 8 μg of SN2 protein is loaded in each lane. Right panel, Cdc45 IP.
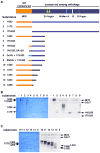
Figure 3. In vitro phosphorylation of Mcm4 fragments using purified DDK
(A) Above, domain structure of Mcm4. Below, protein substrates for the kinase assay. Amino acid residues of each substrate is indicated. Substrate 7 has 7 S/T to A mutations. Substrate 8 has 3xHA tag replacing the NSD. Substrate 9 is derived from substrate 8 by mutating some of the HA sequence to phospho-acceptors. See Fig. 6A for N-terminal sequences of substrates 6–9. (B) Kinase assay of substrates 1–9 in (A). Left, Coomassie Blue-stained gel of the kinase assay. Right, autoradiography of the same gel. Each lane presents 10 μl kinase reaction using 15 nM kinase and 0.5 μM substrate. The first lane (−) has no substrate. (C) Kinase assay of substrates 10–14 in (A). Same condition as in (B).
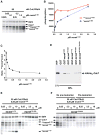
Figure 4. Processive phosphorylation of mcm41-333 through a kinase docking mechanism
(A) Phosphorylation of mcm41-333 as a function of mcm41-333 concentrations. 10 μl of kinase reaction using indicated concentrations of kinase and substrate is in each lane. ***mcm41-333 and *mcm41-333, hyper-phosphorylated and hypo-phosphorylated products, respectively. Auto-phosphorylated Cdc7 and Dbf4 are indicated. (B) Quantification of hyper-phosphorylated and hypo-phosphorylated forms of mcm41-333 in (A) using Phospho Imager. The Y-axis unit is arbitrary. (C) The ratio of hyper-phosphorylated over hypo-phosphorylated mcm41-333 as a function of mcm41-333 concentrations calculated from (B). (D) GST-pull down of purified DDK by GST-mcm4 fusions. Immuno-blot analysis of HAHis6-Cdc7 using 12CA5 antibody. 5 % of the input DDK (INP) and 30 % of pull-down samples using indicated GST fusions as bait were analyzed. (E) Mcm4175-333 added in trans can interfere with phosphorylation of mcm41-333. Each lane presents 10 μl of kinase reaction using indicated amount of DDK, mcm41-333 and mcm41-175 or mcm4175-333 as competitors. Labels are the same as in (A). (F) Pre-incubation of DDK and mcm41-333 eliminates the inhibitory effect of mcm4175-333. No Pre-incubation, same conditions as in (E). Pre-incubation, DDK and mcm41-333 were pre-incubated at 30oC for 10 minutes and then on ice for 1 hour before addition of the mcm4175-333 fragment.
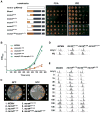
Figure 5. NSD is important for normal cell growth and S-phase progression
(A) Complementation of mcm4Δ by empty vector or plasmids carrying MCM4 alleles with full-length NSD, truncated NSD, or NSD replaced by the N-terminus of Mcm2 using plasmid shuffle assay. (B) Growth curve of cells carrying MCM4, mcm4Δ2-174, mcm4Δ2-145 or mcm21-200-mcm4Δ2-174 as the sole copy of MCM4 on pRS415. (C) The cold-sensitive phenotype of mcm4Δ2-174 cells. Cells with indicated MCM alleles were streaked on YPD plates and incubated at 25oC for 2 days and 16o C for 10 days. (D) mcm4Δ2-174 cells accumulate in S phase. Flow cytometry analysis of DNA content in asynchronous cells in log phase and 4 hours after HU arrest. (E) Cell cycle progression of MCM4, mcm4Δ2-174 and mcm21-200-mcm4Δ2-175 cells. Cells were arrested in G1 by α-factor, released into fresh YPD medium at room temperature, collected at indicated time points, analyzed for DNA content by flow cytometry.
Figure 6. DDK phosphorylation sites at the N-terminus of Mcm4 are important in vivo
(A) Sequences of mcm4 mutants with altered N-terminus. (B) Complementation of mcm4Δ by plasmids carrying mcm4 alleles described in (A). (C) Cells with indicated MCM alleles were streaked on YPD plates and incubated at 25oC and 15oC for 7 days.
Figure 7. Phosphorylation of MCM by DDK may be important for proper engagement of Cdc45 to the MCM complex
(A) Overproduction of Cdc45 is detrimental in NSD mutants of MCM4. Cells with indicated MCM alleles carrying control vector (pVF1) or GAL1,10::CDC45 (p_GAL::CDC45_) were streaked on an SC-URA plate (left) or an SC-URA-Gal plate (right) and incubated at 30oC for 4 days. (B) A model for the recruitment of DDK to licensed origins to regulate proper assembly of pre-IC components during S phase. The ordered recruitment of Cdc45, Sld3 and GINS is largely based on a recent publication (Kanemaki and Labib, 2006). Other factors are omitted for simplicity.
Similar articles
- The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4.
Sheu YJ, Stillman B. Sheu YJ, et al. Nature. 2010 Jan 7;463(7277):113-7. doi: 10.1038/nature08647. Nature. 2010. PMID: 20054399 Free PMC article. - Dbf4 and Cdc7 proteins promote DNA replication through interactions with distinct Mcm2-7 protein subunits.
Ramer MD, Suman ES, Richter H, Stanger K, Spranger M, Bieberstein N, Duncker BP. Ramer MD, et al. J Biol Chem. 2013 May 24;288(21):14926-35. doi: 10.1074/jbc.M112.392910. Epub 2013 Apr 2. J Biol Chem. 2013. PMID: 23549044 Free PMC article. - Structural changes in Mcm5 protein bypass Cdc7-Dbf4 function and reduce replication origin efficiency in Saccharomyces cerevisiae.
Hoang ML, Leon RP, Pessoa-Brandao L, Hunt S, Raghuraman MK, Fangman WL, Brewer BJ, Sclafani RA. Hoang ML, et al. Mol Cell Biol. 2007 Nov;27(21):7594-602. doi: 10.1128/MCB.00997-07. Epub 2007 Aug 27. Mol Cell Biol. 2007. PMID: 17724082 Free PMC article. - Initiating DNA synthesis: from recruiting to activating the MCM complex.
Lei M, Tye BK. Lei M, et al. J Cell Sci. 2001 Apr;114(Pt 8):1447-54. doi: 10.1242/jcs.114.8.1447. J Cell Sci. 2001. PMID: 11282021 Review. - DDK promotes DNA replication initiation: Mechanistic and structural insights.
Li N, Gao N, Zhai Y. Li N, et al. Curr Opin Struct Biol. 2023 Feb;78:102504. doi: 10.1016/j.sbi.2022.102504. Epub 2022 Dec 14. Curr Opin Struct Biol. 2023. PMID: 36525878 Review.
Cited by
- ATR-mediated phosphorylation of FANCI regulates dormant origin firing in response to replication stress.
Chen YH, Jones MJ, Yin Y, Crist SB, Colnaghi L, Sims RJ 3rd, Rothenberg E, Jallepalli PV, Huang TT. Chen YH, et al. Mol Cell. 2015 Apr 16;58(2):323-38. doi: 10.1016/j.molcel.2015.02.031. Epub 2015 Apr 2. Mol Cell. 2015. PMID: 25843623 Free PMC article. - Cryo-EM structure of human hexameric MCM2-7 complex.
Xu N, Lin Q, Tian H, Liu C, Wang P, Suen CM, Yang H, Xiang Y, Zhu G. Xu N, et al. iScience. 2022 Aug 17;25(9):104976. doi: 10.1016/j.isci.2022.104976. eCollection 2022 Sep 16. iScience. 2022. PMID: 36117988 Free PMC article. - A structural framework for replication origin opening by AAA+ initiation factors.
Duderstadt KE, Berger JM. Duderstadt KE, et al. Curr Opin Struct Biol. 2013 Feb;23(1):144-53. doi: 10.1016/j.sbi.2012.11.012. Epub 2012 Dec 21. Curr Opin Struct Biol. 2013. PMID: 23266000 Free PMC article. Review. - Interactions of the human MCM-BP protein with MCM complex components and Dbf4.
Nguyen T, Jagannathan M, Shire K, Frappier L. Nguyen T, et al. PLoS One. 2012;7(4):e35931. doi: 10.1371/journal.pone.0035931. Epub 2012 Apr 23. PLoS One. 2012. PMID: 22540012 Free PMC article. - Claspin recruits Cdc7 kinase for initiation of DNA replication in human cells.
Yang CC, Suzuki M, Yamakawa S, Uno S, Ishii A, Yamazaki S, Fukatsu R, Fujisawa R, Sakimura K, Tsurimoto T, Masai H. Yang CC, et al. Nat Commun. 2016 Jul 12;7:12135. doi: 10.1038/ncomms12135. Nat Commun. 2016. PMID: 27401717 Free PMC article.
References
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. - PubMed
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials