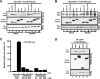RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2 - PubMed (original) (raw)
RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2
Akihiko Komuro et al. J Virol. 2006 Dec.
Abstract
Antiviral innate immune responses can be triggered by accumulation of intracellular nucleic acids resulting from virus infections. Double-stranded RNA (dsRNA) can be detected by the cytoplasmic RNA helicase proteins RIG-I and MDA5, two proteins that share sequence similarities within a caspase recruitment domain (CARD) and a DExD/H box RNA helicase domain. These proteins are considered dsRNA sensors and are thought to transmit the signal to the mitochondrial adapter, IPS-1 (also known as MAVS, VISA, or CARDIF) via CARD interactions. IPS-1 coordinates the activity of protein kinases that activate transcription factors needed to induce beta interferon (IFN-beta) gene transcription. Another helicase protein, LGP2, lacks the CARD region and does not activate IFN-beta gene expression. LGP2 mRNA is induced by interferon, dsRNA treatments, or Sendai virus infection and acts as a feedback inhibitor for antiviral signaling. Results indicate that LGP2 can inhibit antiviral signaling independently of dsRNA or virus infection intermediates by engaging in a protein complex with IPS-1. Experiments suggest that LGP2 can compete with the kinase IKKi (also known as IKKepsilon) for a common interaction site on IPS-1. These results provide the first demonstration of protein interaction as an element of negative-feedback regulation of intracellular antiviral signaling by LGP2.
Figures
FIG. 1.
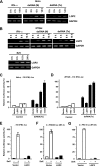
LGP2 is a feedback inhibitor of the antiviral system. (A and B) HeLa (A) or 2fTGH (B) cells were treated with 1,000 U/ml IFN-α or poly(I:C) {both extracellular [dsRNA (M)] and intracellular [dsRNA (Tx)] } or infected with Sendai virus (MOI = 1) for the indicated time course. Total RNA was isolated from cells and subjected to reverse transcriptase (RT)-PCR with the gene-specific primer sets indicated. (C and D) HeLa (C) or 2fTGH (D) cells were transiently transfected with the −110 IFNβ-luc reporter gene and the indicated RNA helicase vector. The cells were mock treated (control) or transfected with poly(I:C) for 6 h [dsRNA (Tx)]. (E) 2fTGH cells transfected with the LGP2 expression vector or empty vector (vector) along with the −110 IFNβ-luc reporter gene were mock treated or infected with Sendai virus (MOI = 1) for 12 h. (F) Similar analysis as for panel E was carried out with the 3× PRD III/I luc reporter gene. (G) Similar analysis as for panel E was carried out with the 4× PRDII luc reporter gene. In all cases, the data were normalized to results for the cotransfected Renilla luciferase reporter and the bars indicate the average (n = 3) ± the standard deviation, plotted as the percent control for each reporter gene. SeV, Sendai virus.
FIG. 2.
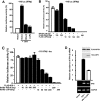
LGP2 negatively regulates endogenous antiviral gene expression. (A to C) Effect of LGP2 interference on antiviral response gene induction. 2fTGH cells were transfected with siRNA for LGP2, RIG-I, or MDA5 (as indicated in inset boxes), and total RNA was processed for real time RT-PCR. The relative abundances of IFN-β mRNA (A), RIG-I mRNA (B), and MDA5 mRNA (C) were quantified by real time RT-PCR. The Student's t test was used for statistical analysis, and P values are indicated. These results were representative of at least two independent transfection experiments. Error bars indicate the standard deviations for the results of duplicate PCRs. N.D., not detected. (D) Demonstration of the efficacy of siRNA-mediated LGP2 knockdown. 2fTGH cells were transfected with a control scrambled siRNA duplex or LGP2-specific siRNA and then infected with Sendai virus or mock infected. Total RNA was prepared and subjected to RT-PCR amplification with LGP2- or GAPDH-specific primers. (E) 2fTGH cells transfected with empty vector (+ Vector) or LGP2 expression vector (+ LGP2) were infected with Sendai virus for the indicated time course. Total RNA was isolated from cells and subjected to RT-PCR with the gene-specific primer sets indicated. SeV, Sendai virus.
FIG. 3.
LGP2 inhibits RIG-I signaling independently of dsRNA or virus infection. (A) HEK 293T cells were transfected with 50 ng of the −110 IFNβ-luc reporter plasmid along with 100 ng of the expression vector for RIG-IC or LGP2. Twenty-four hours after being transfected, the cells were infected with Sendai virus (SeV) for 8 h. Lysates were prepared and subjected to luciferase assays, plotted as the percent positive control. (B) HEK 293T cells were transfected with 50 ng of the −110 IFNβ-luc reporter plasmid along with 150 ng of the expression vector for RIG-IN alone or with increasing amounts of the expression plasmid for LGP2 or RIG-IC, as indicated. The lysates were prepared and subjected to luciferase assays. (C) HEK 293T cells were transfected with 50 ng of the −110 IFNβ-luc reporter plasmid along with 100 ng of the expression vector for IPS-1 alone or with increasing amounts of the expression plasmid for LGP2 or RIG-IC, as indicated. (D) HEK 293T cells were transfected with the expression vector for FLAG-tagged IPS-1 alone or along with the expression vector for HA-tagged LGP2. After 24 h, total RNA was isolated for quantitative real time RT-PCR (top graph) or nonquantitative RT-PCR followed by agarose gel electrophoresis and ethidium bromide staining (bottom). The relative abundance of IFN-β mRNA was normalized to the abundance of GAPDH mRNA. The expression of proteins was confirmed by immunoblotting (inset). The data are representative of at least two independent transfection experiments. The error bars indicate the standard deviation for duplicate PCRs.
FIG. 4.
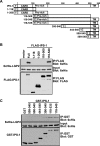
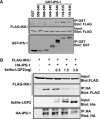
LGP2 coprecipitates with IPS-1 and RIG-I. (A) HEK 293T cells were transfected with the expression vector for Myc-tagged RIG-I and the expression vector for FLAG-tagged IPS-1 along with the expression vector for His-tagged LGP2 or His-tagged Yap. Cell lysates were subjected to FLAG immunoprecipitation using M2-agarose beads and immunoblotting for Myc or His tags. (B) HEK 293T cells were transfected with the expression vector for Myc-tagged RIG-I and the expression vector for FLAG-tagged IPS-1 along with increasing amounts of the expression vector for His-tagged LGP2. Lysates were subjected to FLAG immunoprecipitation as for panel A. (C) HEK 293T cells were transfected with the expression vector for His-tagged RIG-IC or LGP2 along with the expression vector for FLAG-tagged IPS-1 or empty vector. Lysates were subjected to FLAG immunoprecipitation followed by immunoblotting, using His-tag antibody. (D) Lysates from Sendai virus-infected HEK 293T cells were subjected to immunoprecipitation with LGP2 antibody or control rabbit IgG. The precipitated proteins were analyzed by anti-Cardif antibody, and the same blot was reprobed with LGP2 antibody.
FIG. 5.
LGP2 coprecipitates RIG-I and MDA5 following virus infection. (A) HEK 293T cells were transfected with expression vectors for HA-tagged LGP2 and FLAG-tagged LGP2, RIG-I, or MDA5. The cells were infected with Sendai virus or mock infected for 6 h. Cell lysates were subjected to FLAG immunoprecipitation using M2-agarose beads. The HA-LGP2 proteins copurified with FLAG-tagged RNA helicases were analyzed by immunoblotting, using HA antibody (Roche). (B) Similar to procedure for panel A but using Myc-tagged RIG-I along with FLAG-tagged LGP2, LGP2 mutant (K30A), RIG-I, or MDA5. FLAG eluates were analyzed by immunoblotting using Myc antibody (Roche). An asterisk indicates a nonspecific band, and an arrowhead indicates Myc-RIG-I. (C) The LGP2 Walker A motif mutation K30A retains inhibitory activity. A luciferase assay was carried out as for Fig. 3A, comparing the activities of wild-type (WT) and mutant (K30A) LGP2 resulting from transfection of increasing amounts of expression vectors from 20 to 600 ng, as indicated. (D) HEK 293T cells were transfected with expression vectors for HA-tagged LGP2 and FLAG-tagged LGP2, RIG-I, RIG-IN, or RIG-IC and infected with Sendai virus for 6 h. Lysates were subjected to FLAG immunoprecipitation as in for panel A. Coprecipitated LGP2 protein was detected by immunoblotting, using HA antibody (Roche).
FIG. 6.
LGP2 interaction requires the C-terminal and the transmembrane regions of IPS-1. (A) Schematic representation of IPS-1 fragments used for mapping. Positions of FLAG or GST tags (T), CARD, the proline-rich region (Pro-rich), and the transmembrane domain (TM) are indicated. (B) HEK 293T cells were transfected with expression vectors for six-His-tagged LGP2 and the FLAG-tagged regions of IPS-1, as indicated. Cell lysates were subjected to FLAG immunoprecipitation using M2-agarose beads. His-LGP2 protein copurification with FLAG-tagged IPS-1 was analyzed by immunoblotting, using six-His antibody. (C) HEK 293T cells were transfected with expression vectors for His-tagged LGP2 and GST-tagged regions of IPS-1, as indicated. Cell lysates were subjected to glutathione bead affinity purification. Six-His-LGP2 protein copurification with GST-IPS-1 was analyzed by immunoblotting, using six-His antibody.
FIG. 7.
LGP2 competes with IKKi for IPS-1binding. (A) HEK 293T cells were transfected with expression vectors for FLAG-tagged IKKi and GST-IPS-1 fusions, as indicated. Cell lysates were subjected to glutathione bead affinity purification. FLAG-tagged IKKi copurification with GST-IPS-1 was analyzed by immunoblotting, using FLAG antibody. (B) HEK 293T cells were transfected with expression vectors for FLAG-tagged IKKi and HA-tagged IPS-1 along with increasing amounts of the expression vector for six-His-tagged LGP2. Cell lysates were subjected to HA purification. FLAG-tagged IKKi purified with HA-IPS-1 was analyzed by immunoblotting, using FLAG antibody.
FIG. 8.
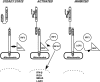
Model of LGP2 action in dsRNA- and virus-independent inhibition of antiviral signal transduction. Schematic diagrams are drawn to illustrate key features of the results and interpretation. For simplicity, a single CARD-helicase protein is illustrated. Many components of the antiviral response and additional unknown components that might bridge indirect protein interactions are not depicted. (A) In the steady state, the absence of virus infection or cytosolic dsRNA maintains the CARD-helicase RNA sensor protein in an autoinhibited quiescent state. IPS-1 is anchored in the mitochondria, where it may associate with IKKi. IRF3 is unphosphorylated and in its latent cytoplasmic location. (B) Once activated by virus infection or cytosolic dsRNA, the CARD of the RNA sensor is activated to make heterotypic contact with the CARD of IPS-1. This activates IKKi to phosphorylate IRF3, inducing its nuclear translocation and transcriptional activation of target genes, including IFN-β, RIG-I, MDA5, and LGP2. (C) LGP2 accumulation in the cell leads to signal inhibition. LGP2 interaction displaces IKKi from the IPS-1 C terminus to disengage signal propagation. In this case, the CARD-helicase can remain in association with the IPS-1 plus LGP2 complexes.
Similar articles
- Regulation of interferon production by RIG-I and LGP2: a lesson in self-control.
Vitour D, Meurs EF. Vitour D, et al. Sci STKE. 2007 May 1;2007(384):pe20. doi: 10.1126/stke.3842007pe20. Sci STKE. 2007. PMID: 17473309 - Paramyxovirus V protein interaction with the antiviral sensor LGP2 disrupts MDA5 signaling enhancement but is not relevant to LGP2-mediated RLR signaling inhibition.
Rodriguez KR, Horvath CM. Rodriguez KR, et al. J Virol. 2014 Jul;88(14):8180-8. doi: 10.1128/JVI.00737-14. Epub 2014 May 14. J Virol. 2014. PMID: 24829334 Free PMC article. - LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses.
Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. Satoh T, et al. Proc Natl Acad Sci U S A. 2010 Jan 26;107(4):1512-7. doi: 10.1073/pnas.0912986107. Epub 2010 Jan 8. Proc Natl Acad Sci U S A. 2010. PMID: 20080593 Free PMC article. - LGP2 synergy with MDA5 in RLR-mediated RNA recognition and antiviral signaling.
Bruns AM, Horvath CM. Bruns AM, et al. Cytokine. 2015 Aug;74(2):198-206. doi: 10.1016/j.cyto.2015.02.010. Epub 2015 Mar 18. Cytokine. 2015. PMID: 25794939 Free PMC article. Review. - Regulation of RLR-mediated innate immune signaling--it is all about keeping the balance.
Eisenächer K, Krug A. Eisenächer K, et al. Eur J Cell Biol. 2012 Jan;91(1):36-47. doi: 10.1016/j.ejcb.2011.01.011. Epub 2011 Apr 9. Eur J Cell Biol. 2012. PMID: 21481967 Review.
Cited by
- Cancer therapies activate RIG-I-like receptor pathway through endogenous non-coding RNAs.
Ranoa DR, Parekh AD, Pitroda SP, Huang X, Darga T, Wong AC, Huang L, Andrade J, Staley JP, Satoh T, Akira S, Weichselbaum RR, Khodarev NN. Ranoa DR, et al. Oncotarget. 2016 May 3;7(18):26496-515. doi: 10.18632/oncotarget.8420. Oncotarget. 2016. PMID: 27034163 Free PMC article. - Interplay between interferon-mediated innate immunity and porcine reproductive and respiratory syndrome virus.
Sun Y, Han M, Kim C, Calvert JG, Yoo D. Sun Y, et al. Viruses. 2012 Apr;4(4):424-46. doi: 10.3390/v4040424. Epub 2012 Apr 2. Viruses. 2012. PMID: 22590680 Free PMC article. Review. - Antiviral RNA recognition and assembly by RLR family innate immune sensors.
Bruns AM, Horvath CM. Bruns AM, et al. Cytokine Growth Factor Rev. 2014 Oct;25(5):507-12. doi: 10.1016/j.cytogfr.2014.07.006. Epub 2014 Jul 15. Cytokine Growth Factor Rev. 2014. PMID: 25081315 Free PMC article. Review. - Regulation of RIG-I-like receptor-mediated signaling: interaction between host and viral factors.
Onomoto K, Onoguchi K, Yoneyama M. Onomoto K, et al. Cell Mol Immunol. 2021 Mar;18(3):539-555. doi: 10.1038/s41423-020-00602-7. Epub 2021 Jan 18. Cell Mol Immunol. 2021. PMID: 33462384 Free PMC article. Review. - NLRs, inflammasomes, and viral infection.
Jacobs SR, Damania B. Jacobs SR, et al. J Leukoc Biol. 2012 Sep;92(3):469-77. doi: 10.1189/jlb.0312132. Epub 2012 May 10. J Leukoc Biol. 2012. PMID: 22581934 Free PMC article. Review.
References
- Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124:783-801. - PubMed
- Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKɛ and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. - PubMed
- Gresser, I., M. G. Tovey, C. Maury, and I. Chouroulinkov. 1975. Lethality of interferon preparations for newborn mice. Nature 258:76-78. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI048722/AI/NIAID NIH HHS/United States
- R21 AI048722/AI/NIAID NIH HHS/United States
- R01 GM063872/GM/NIGMS NIH HHS/United States
- GM063872/GM/NIGMS NIH HHS/United States
- AI048722/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous