Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures - PubMed (original) (raw)
Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures
Patrick D Schloss et al. Appl Environ Microbiol. 2006 Oct.
Abstract
The recent advent of tools enabling statistical inferences to be drawn from comparisons of microbial communities has enabled the focus of microbial ecology to move from characterizing biodiversity to describing the distribution of that biodiversity. Although statistical tools have been developed to compare community structures across a phylogenetic tree, we lack tools to compare the memberships and structures of two communities at a particular operational taxonomic unit (OTU) definition. Furthermore, current tests of community structure do not indicate the similarity of the communities but only report the probability of a statistical hypothesis. Here we present a computer program, SONS, which implements nonparametric estimators for the fraction and richness of OTUs shared between two communities.
Figures
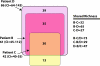
FIG. 1.
Venn diagram comparing the OTU0.03 memberships found in the distal esophagi of three patients (patient B[n = 205], patient C [n = 264], and patient D [n = 245]). Below each patient's name is the Chao1 richness estimate and the 95% confidence interval for that community. We estimated the richness of the overlapping regions based on the pairwise _S_1,2 Chao richness estimates shared by the three communities and by pooling two communities and estimating the fraction shared with the third community. These estimates are provided on the right side of the figure. The Chao1 richness estimate of the three libraries pooled together was 125 (CI = 98 to 191), and the sum of the individual sectors in the diagram was 117.
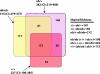
FIG. 2.
Venn diagram comparing the pooled OTU0.03 memberships found in the gastrointestinal tracts and stool samples of three patients (patient 70 [n = 4,392], patient 71 [n = 3,605], and patient 72 [n = 3,834]). The Chao1 richness estimate of the three libraries pooled together was 469 (CI = 425 to 544), and the sum of the individual sectors in the diagram was 433.
FIG. 3.
Unweighted pair group method with arithmetic mean dendrogram comparing the pairwise θ values between the seven gastrointestinal tract tissue and stool specimens sampled from three patients. The length of the reference bar represents a distance of 0.10 (distance = 1 − θ). “>” indicates that the calculated richness value represents a minimum because the estimate did not stabilize with respect to sampling effort.
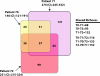
FIG. 4.
Venn diagram comparing the OTU0.03 memberships found among pooled +/+ (n = 876), ob/+ (n = 1,186), and ob/ob (n = 1,327) mice. The Chao1 richness estimate of the three libraries pooled together was 623 (CI = 535 to 761), and the sum of the individual sectors in the diagram was 554.
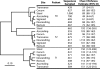
FIG. 5.
Unweighted pair group method with arithmetic mean dendrogram comparing nonparametric estimates of the abundance-based Jaccard similarity coefficients for the OTU0.03s identified among 19 mouse cecum samples. A dendrogram constructed using the estimated Sørenson similarity index had the same topology. Although the exact branching orders were not identical, in the θ dendrogram, the OTU0.03 community structures of the offspring generally clustered with the mother; the community structures of mice M2A-3 and M2B-1 clustered with mother 3. The community name corresponds to the mother (e.g., M1-1 is the offspring of mother 1). The length of the reference bar represents a distance of 0.10 (distance = 1 − _J_abund). “>” indicates that the calculated richness value represents a minimum because the estimate did not stabilize with respect to sampling effort.
References
- Anderson, M. J. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26:32-46.
- Burnham, K. P., and W. S. Overton. 1979. Robust estimation of population size when capture probabilities vary among animals. Ecology 60:927-936.
- Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270.
- Chao, A., R. L. Chazdon, R. K. Colwell, and T.-J. Shen. 2006. Abundance-based similarity indices and their estimation when there are unseen species in samples. Biometrics 62:361-371. - PubMed
- Chao, A., R. L. Chazdon, R. K. Colwell, and T. J. Shen. 2005. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol. Lett. 8:148-159.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources