Possible errors in the interpretation of ethidium bromide and PicoGreen DNA staining results from ethidium monoazide-treated DNA - PubMed (original) (raw)
Comment
Possible errors in the interpretation of ethidium bromide and PicoGreen DNA staining results from ethidium monoazide-treated DNA
Ingeborg Hein et al. Appl Environ Microbiol. 2006 Oct.
No abstract available
Figures
FIG.1.
Analysis of EMA-treated and EMA-untreated DNA on an ethidium bromide-stained agarose gel. EMA treatment was performed, as published recently, in a volume of 1 ml at a concentration of 100 μg EMA (Molecular Probes, Eugene, OR)/ml (3). The samples were incubated for 5 min at 4°C in the dark, followed by exposure to light in macrocuvettes (Greiner, Frickenhausen, Germany) for 2 min at a distance of 20 cm from the light source, using a Ventilux 1250 lamp and a 650-W halogen light bulb (Hedler, Runkel, Germany). Calf thymus DNA was obtained from Sigma-Aldrich (St. Louis, MO). Salmonella enterica serovar Typhimurium NCTC 12023 DNA was isolated from an overnight culture in tryptic soy broth with 0.6% yeast at 37°C using the NucleoSpin tissue kit (Machery-Nagel, Düren, Germany). Lane m, 100-bp ladders. Lanes marked with a contain samples of DNA exposed (exp.) to light without the addition of EMA.
FIG.1.
Lysis matrix tubes with pelleted cell debris of viable and dead Salmonella and E. coli O157:H7 without (−) or with (+) previous EMA treatment. Culture aliquots were either processed directly without stress exposure, heat treated at 72°C for 15 min, or exposed to 70% isopropanol (isoprop.) for 10 min. Cells were pelleted by centrifugation. Cell pellets were resuspended and subjected to bead beating. Lysis matrix tubes are shown after centrifugation for 5 min at 14,000 × g, with the cell debris facing upward.
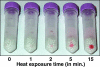
FIG. 2.
Lysis matrix tubes with pelleted cell debris of Salmonella exposed to heat stress at 72°C (1 to 15 min) with subsequent EMA treatment. Cells were pelleted by centrifugation. Cell pellets were resuspended and subjected to bead beating. Lysis matrix tubes are shown after centrifugation for 5 min at 14,000 × g, with the cell debris facing upward.
Comment on
- Selective removal of DNA from dead cells of mixed bacterial communities by use of ethidium monoazide.
Nocker A, Camper AK. Nocker A, et al. Appl Environ Microbiol. 2006 Mar;72(3):1997-2004. doi: 10.1128/AEM.72.3.1997-2004.2006. Appl Environ Microbiol. 2006. PMID: 16517648 Free PMC article.
Similar articles
- Staining Nucleic Acids.
Green MR, Sambrook J. Green MR, et al. Cold Spring Harb Protoc. 2020 Feb 3;2020(2):098228. doi: 10.1101/pdb.top098228. Cold Spring Harb Protoc. 2020. PMID: 32015006 - Binding of ethidium monoazide to the chromatin in human lymphocytes.
Cantrell CE, Yielding KL. Cantrell CE, et al. Biochim Biophys Acta. 1980 Aug 26;609(1):173-9. doi: 10.1016/0005-2787(80)90210-5. Biochim Biophys Acta. 1980. PMID: 6157417 - Quantifying DNA concentrations using fluorometry: a comparison of fluorophores.
Rengarajan K, Cristol SM, Mehta M, Nickerson JM. Rengarajan K, et al. Mol Vis. 2002 Nov 6;8:416-21. Mol Vis. 2002. PMID: 12432341 - Electroanalytical study of SYBR Green I and ethidium bromide intercalation in methylated and unmethylated amplicons.
Ioannou AK, Alexiadou DK, Kouidou SA, Voulgaropoulos AN, Girousi ST. Ioannou AK, et al. Anal Chim Acta. 2010 Jan 11;657(2):163-8. doi: 10.1016/j.aca.2009.10.047. Anal Chim Acta. 2010. PMID: 20005328 - Bioconjugation via azide-Staudinger ligation: an overview.
Schilling CI, Jung N, Biskup M, Schepers U, Bräse S. Schilling CI, et al. Chem Soc Rev. 2011 Sep;40(9):4840-71. doi: 10.1039/c0cs00123f. Epub 2011 Jun 17. Chem Soc Rev. 2011. PMID: 21687844 Review.
Cited by
- Monochloramine disinfection kinetics of Nitrosomonas europaea by propidium monoazide quantitative PCR and Live/dead BacLight methods.
Wahman DG, Wulfeck-Kleier KA, Pressman JG. Wahman DG, et al. Appl Environ Microbiol. 2009 Sep;75(17):5555-62. doi: 10.1128/AEM.00407-09. Epub 2009 Jun 26. Appl Environ Microbiol. 2009. PMID: 19561179 Free PMC article. - Removal of free extracellular DNA from environmental samples by ethidium monoazide and propidium monoazide.
Wagner AO, Malin C, Knapp BA, Illmer P. Wagner AO, et al. Appl Environ Microbiol. 2008 Apr;74(8):2537-9. doi: 10.1128/AEM.02288-07. Epub 2008 Feb 22. Appl Environ Microbiol. 2008. PMID: 18296534 Free PMC article. - New perspectives on viable microbial communities in low-biomass cleanroom environments.
Vaishampayan P, Probst AJ, La Duc MT, Bargoma E, Benardini JN, Andersen GL, Venkateswaran K. Vaishampayan P, et al. ISME J. 2013 Feb;7(2):312-24. doi: 10.1038/ismej.2012.114. Epub 2012 Oct 11. ISME J. 2013. PMID: 23051695 Free PMC article. - Use of high throughput amplicon sequencing and ethidium monoazide dye to track microbiota changes in an equol-producing menopausal woman receiving a long-term isoflavones treatment.
Guadamuro L, Azcárate-Peril MA, Tojo R, Mayo B, Delgado S. Guadamuro L, et al. AIMS Microbiol. 2019 Mar 22;5(1):102-116. doi: 10.3934/microbiol.2019.1.102. eCollection 2019. AIMS Microbiol. 2019. PMID: 31384706 Free PMC article.
References
- Nogva, H. K., S. M. Drømtørp, H. Nissen, and K. Rudi. 2003. Ethidium monoazide for DNA-based differentiation of viable and dead bacteria by 5′-nuclease PCR. BioTechniques 34:804-813. - PubMed
- Rudi, K., H. K. Nogva, B. Moen, H. Nissen, S. Bredholt, T. Møretrø, K. Naterstad, and A. Holck. 2002. Development and application of new nucleic acid-based technologies for microbial community analyses in foods. Int. J. Food Microbiol. 78:171-180. - PubMed
- Yielding, L. W., and W. J. Firth III. 1980. Structure-function characterization for ethidium photoaffinity labels as mutagens in Salmonella. Mutat. Res. 71:161-168. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources