The WD repeat-containing protein IFTA-1 is required for retrograde intraflagellar transport - PubMed (original) (raw)
The WD repeat-containing protein IFTA-1 is required for retrograde intraflagellar transport
Oliver E Blacque et al. Mol Biol Cell. 2006 Dec.
Abstract
The assembly and maintenance of cilia require intraflagellar transport (IFT), a microtubule-dependent bidirectional motility of multisubunit protein complexes along ciliary axonemes. Defects in IFT and the functions of motile or sensory cilia are associated with numerous human ailments, including polycystic kidney disease and Bardet-Biedl syndrome. Here, we identify a novel Caenorhabditis elegans IFT gene, IFT-associated gene 1 (ifta-1), which encodes a WD repeat-containing protein with strong homology to a mammalian protein of unknown function. Both the C. elegans and human IFTA-1 proteins localize to the base of cilia, and in C. elegans, IFTA-1 can be observed to undergo IFT. IFTA-1 is required for the function and assembly of cilia, because a C. elegans ifta-1 mutant displays chemosensory abnormalities and shortened cilia with prominent ciliary accumulations of core IFT machinery components that are indicative of retrograde transport defects. Analyses of C. elegans IFTA-1 localization/motility along bbs mutant cilia, where anterograde IFT assemblies are destabilized, and in a che-11 IFT gene mutant, demonstrate that IFTA-1 is closely associated with the IFT particle A subcomplex, which is implicated in retrograde IFT. Together, our data indicate that IFTA-1 is a novel IFT protein that is required for retrograde transport along ciliary axonemes.
Figures
Figure 1.
ifta-1 (C54G7.4) is an X-box–containing gene that is conserved in ciliated organisms. (A) RFX transcription factor-binding X-boxes are found upstream of ifta-1 in C. elegans, C. briggsae, and H. sapiens. (B) Schematic of the IFTA-1 protein, which consists of 1199 amino acids (in C. elegans) and is predicted to possess three WD protein motifs. (C) Cross-species comparison of the percentage of amino acid identity between IFTA-1 homologues in C. elegans, H. sapiens, Mus musculus, pufferfish, Drosophila melanogaster, C. reinhardtii, Trypanosoma brucei, and Paramecium. Note that IFTA-1 homologues are not present in nonciliated organisms such as A. thaliana and S. cerevisiae.
Figure 2.
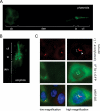
IFTA-1 localizes to ciliary structures in C. elegans and to the basal body/centrosomal region of mammalian kidney cells. (A and B) GFP-tagged IFTA-1 localizes specifically at the base of cilia (transition zones or basal bodies) and along the ciliary axonemes of C. elegans ciliated neurons. Shown are fluorescence images of one set of phasmid neurons in the nematode tail (A), and one set of amphid cilia in nematode head (B), with the cell bodies (cb), dendrites (den), transition zones (tz), and ciliary axonemes (cil) indicated. (C) GFP-tagged H. sapiens IFTA-1 localizes at the basal bodies and centrosomes of mammalian kidney cells. Shown are coimmunofluorescence images of ICMD3 cells expressing an H. sapiens ifta1::gfp transgene. Top, antibodies to endogenous γ-tubulin and acetylated α-tubulin detect centrosomes/basal bodies (red, bracket) and ciliary axonemes (red, arrowhead), respectively. Middle, GFP-tagged IFTA1 displays punctate fluorescence (brackets), consistent with its localization at basal bodies and centrosomes. Bottom, merged red and green fluorescence images demonstrate colocalization of basal bodies/centrosomes and GFP-tagged IFTA1 (yellow, bracket).
Figure 3.
C. elegans IFTA-1 undergoes biphasic IFT. (A) GFP-tagged IFTA-1 undergoes IFT. Shown is a kymograph (3 consecutive 0.5-s time points) depicting the anterograde motility (away from the transition zone, tz) of a GFP-tagged IFTA-1–associated particle (arrowhead) along the ciliary axoneme of one phasmid neuron. Scale bar is shown at the bottom of the kymograph. (B and C) The anterograde IFT motility of IFTA-1 is biphasic. Shown in B are representative “still” fluorescence images and corresponding kymographs (M and D) and kymograph schematics (M′ and D′), obtained from the analysis of IFTA-1::GFP transport (using IFT motility assays) along amphid sensory cilia. Note that the individual diagonal lines of the kymographs correspond to motility rates for individual fluorescent particles. Images (B) and table (C) show that in wild-type cilia, GFP-tagged IFTA-1 displays biphasic IFT, with intermediate velocities along middle segments (M and M′) and fast velocities along distal segments (D and D′). Also shown is the monophasic IFT behavior of IFTA-1::GFP along the ciliary axonemes of animals with loss-of-function mutations in a heterotrimeric kinesin-II subunit gene, klp-11, and a homodimeric OSM-3-kinesin subunit gene, osm-3.
Figure 4.
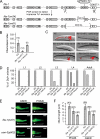
ifta-1 is a ciliogenic gene required for the normal structure and function of sensory cilia. (A) Generation of two mutant alleles of ifta-1 by transposon excision. Shown is the genomic position of cxP5003::Tc1 within ifta-1, and the resultant 600- and 2009-base pair deletions obtained after excision. (B) ifta-1(nx61) mutants are defective in chemotaxis toward iso-amyl alcohol compared with wild-type (N2) animals. Rescue of the chemotaxis defect was observed in ifta-1(nx61) worms expressing a wild-type ifta-1::gfp transgene. (C) ifta-1(nx34) and ifta-1(nx61) animals are defective in their ability to uptake the fluorescent dye DiI. Shown are DIC-fluorescence merged images of the head region of wild-type and ifta-1 mutant worms after a dye-fill assay. Note that the dye-fill defective phenotype (Dyf) of ifta-1 mutants is fully rescued in ifta-1 mutants expressing a wild-type ifta-1::gfp transgene. (D) The Dyf phenotype of ifta-1(nx61) mutants is fully penetrant at all larval and adult stages, whereas ifta-1(nx34) animals have a stage-specific Dyf phenotype. Note that each data point represents the analysis of 50 worms. (E and F) The cilia length of ifta-1(nx61) mutants is shorter than wild-type worms but longer than osm-3(p802) mutants. Presented are fluorescence images (E) and a graph (F) showing the cilia lengths obtained for PHA/B phasmid and ASER amphid neurons, by using srb-6p::gfp and gcy-5p::gfp reporters, respectively. For E: Bar, 2 μm; star, transition zone; bracket, cilia. The n values for F are presented in brackets.
Figure 5.
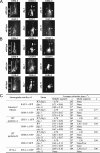
ifta-1 mutants phenocopy the retrograde IFT defects of che-11 mutants. (A and B) The anterograde IFT machinery, namely, the kinesin-2 motors (A) and IFT-B subcomplex proteins (B), accumulates along ifta-1 and che-11 mutant cilia. Shown are representative fluorescence images of one set of amphid cilia from wild-type (N2), ifta-1(nx61), and che-11(e1810) animals, expressing the indicated GFP-tagged protein. Images demonstrate that in contrast to wild-type worms, the anterograde IFT motor subunits (KAP-1::GFP and OSM-3::GFP) and IFT-B subcomplex proteins (OSM-1::GFP, OSM-6::GFP, and CHE-2::GFP) accumulate (see arrowheads) within the ciliary axonemes of ifta-1 and che-11 mutants, indicating a retrograde IFT defect. Note that in wild-type panels, the ciliary axonemes (ax), transition zones (tz), and dendrites (d) are denoted. Note also that all images are similarly sized and orientated, with the transition zone region denoted in all panels by a bracket. (C) Anterograde IFT motility velocities of fluorescent IFT-proteins along the amphid cilia of WT, ifta-1(nx61) and che-11(e1810) mutants. The asterisk denotes that compared with wild-type cilia, the number of detectable anterograde transport events along ifta-1(nx61), and che-11(e1810) mutant cilia was low; hence, only a small n value could be obtained. References (Ref.) refer to Snow et al. (2004) for OSM-6::GFP, Ou et al. (2005a) for KAP-1::GFP, OSM-3::GFP, CHE-2::GFP, and CHE-11::GFP, and Ou and Scholey, unpublished data, for OSM-1::GFP.
Figure 6.
IFTA-1 is an IFT particle subcomplex A-type protein. (A and B) Unlike wild-type (N2) cilia, in bbs mutant cilia, IFTA-1::GFP moves only along the middle segments (M and M′) at slow rates characteristic of uncoupled kinesin-II/IFT-A subcomplex assemblies and does not enter the distal segments. Shown in A are representative still fluorescence images and corresponding kymographs (M) and kymograph schematics (M′), obtained from the analysis of IFTA-1 transport (by using IFT motility assays) along wild-type (N2), bbs-8(nx77), and osm-12(n1606) mutant cilia. All images are similarly sized and orientated, with the transition zones and ciliary axonemes denoted in all panels. (C) CHE-11 protein function is required for the ciliary localization of IFTA-1 but not vice versa. Shown are representative still fluorescence images and corresponding kymographs (M) and kymograph schematics (M′), obtained from the analysis of IFT (by using IFT motility assays) of CHE-11::GFP in ifta-1(nx61) mutants, and IFTA-1::GFP in che-11(e1810) mutants. Note that CHE-11::GFP accumulates (see arrowheads) within the ciliary axonemes of ifta-1 mutants. In contrast, IFTA-1::GFP does not enter the ciliary axonemes of che-11 mutant cilia. Both images are similarly sized and orientated, with the transition zones and ciliary axonemes denoted in both images.
Figure 7.
Model of IFTA-1 function as a component of retrograde IFT. Presented is an extension of previous models by Snow et al. (2004) and Ou et al. (2005a), where two kinesin-2 motors were shown to drive anterograde IFT (heterotrimeric kinesin-II, drawn in brown; and homodimeric OSM-3-kinesin, drawn in blue), and BBS-7 and BBS-8 proteins were found to stabilize the association of IFT-A and IFT-B subcomplexes. (A) Loss of IFTA-1 function causes a ciliary accumulation of IFT machinery components, including IFT-B subcomplexes and the IFT-A subcomplex protein (CHE-11), indicating that IFTA-1 function is not required for entry of IFT machinery assemblies into cilia via anterograde IFT but that it is required for recycling of the IFT machinery back to the base of cilia via retrograde IFT. Defective retrograde IFT results in a partial loss of the singlet-microtubule-derived distal segment. (B) The IFT-A subcomplex gene mutant che-11 closely phenocopies the retrograde IFT defects of ifta-1 mutants. In addition, CHE-11 function is required for the ciliary localization of IFTA-1 protein. (C) Together, the data presented in A and B indicate that in wild-type C. elegans sensory cilia, IFTA-1 is closely associated with IFT-A subcomplexes and functions within the retrograde arm of the IFT process.
Similar articles
- Caenorhabditis elegans DYF-2, an orthologue of human WDR19, is a component of the intraflagellar transport machinery in sensory cilia.
Efimenko E, Blacque OE, Ou G, Haycraft CJ, Yoder BK, Scholey JM, Leroux MR, Swoboda P. Efimenko E, et al. Mol Biol Cell. 2006 Nov;17(11):4801-11. doi: 10.1091/mbc.e06-04-0260. Epub 2006 Sep 6. Mol Biol Cell. 2006. PMID: 16957054 Free PMC article. - The BBSome controls IFT assembly and turnaround in cilia.
Wei Q, Zhang Y, Li Y, Zhang Q, Ling K, Hu J. Wei Q, et al. Nat Cell Biol. 2012 Sep;14(9):950-7. doi: 10.1038/ncb2560. Epub 2012 Aug 26. Nat Cell Biol. 2012. PMID: 22922713 Free PMC article. - Functional genomics of intraflagellar transport-associated proteins in C. elegans.
Inglis PN, Blacque OE, Leroux MR. Inglis PN, et al. Methods Cell Biol. 2009;93:267-304. doi: 10.1016/S0091-679X(08)93014-4. Epub 2009 Dec 4. Methods Cell Biol. 2009. PMID: 20409822 - Intraflagellar transport: mechanisms of motor action, cooperation, and cargo delivery.
Prevo B, Scholey JM, Peterman EJG. Prevo B, et al. FEBS J. 2017 Sep;284(18):2905-2931. doi: 10.1111/febs.14068. Epub 2017 Apr 18. FEBS J. 2017. PMID: 28342295 Free PMC article. Review. - Intraflagellar transport: from molecular characterisation to mechanism.
Blacque OE, Cevik S, Kaplan OI. Blacque OE, et al. Front Biosci. 2008 Jan 1;13:2633-52. doi: 10.2741/2871. Front Biosci. 2008. PMID: 17981739 Review.
Cited by
- Intraflagellar transport and functional analysis of genes required for flagellum formation in trypanosomes.
Absalon S, Blisnick T, Kohl L, Toutirais G, Doré G, Julkowska D, Tavenet A, Bastin P. Absalon S, et al. Mol Biol Cell. 2008 Mar;19(3):929-44. doi: 10.1091/mbc.e07-08-0749. Epub 2007 Dec 19. Mol Biol Cell. 2008. PMID: 18094047 Free PMC article. - Localization of a guanylyl cyclase to chemosensory cilia requires the novel ciliary MYND domain protein DAF-25.
Jensen VL, Bialas NJ, Bishop-Hurley SL, Molday LL, Kida K, Nguyen PA, Blacque OE, Molday RS, Leroux MR, Riddle DL. Jensen VL, et al. PLoS Genet. 2010 Nov 24;6(11):e1001199. doi: 10.1371/journal.pgen.1001199. PLoS Genet. 2010. PMID: 21124868 Free PMC article. - The Trypanosoma brucei flagellum: moving parasites in new directions.
Ralston KS, Kabututu ZP, Melehani JH, Oberholzer M, Hill KL. Ralston KS, et al. Annu Rev Microbiol. 2009;63:335-62. doi: 10.1146/annurev.micro.091208.073353. Annu Rev Microbiol. 2009. PMID: 19575562 Free PMC article. Review. - Regulation of hyperoxia-induced social behaviour in Pristionchus pacificus nematodes requires a novel cilia-mediated environmental input.
Moreno E, Sieriebriennikov B, Witte H, Rödelsperger C, Lightfoot JW, Sommer RJ. Moreno E, et al. Sci Rep. 2017 Dec 14;7(1):17550. doi: 10.1038/s41598-017-18019-0. Sci Rep. 2017. PMID: 29242625 Free PMC article. - Subunit interactions and organization of the Chlamydomonas reinhardtii intraflagellar transport complex A proteins.
Behal RH, Miller MS, Qin H, Lucker BF, Jones A, Cole DG. Behal RH, et al. J Biol Chem. 2012 Apr 6;287(15):11689-703. doi: 10.1074/jbc.M111.287102. Epub 2011 Dec 14. J Biol Chem. 2012. PMID: 22170070 Free PMC article.
References
- Ansley S. J., et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–633. - PubMed
- Avidor-Reiss T., Maer A. M., Koundakjian E., Polyanovsky A., Keil T., Subramaniam S., Zuker C. S. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. - PubMed
- Badano J. L., Mitsuma N., Beales P. L., Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 2006 [Epub ahead of print] - PubMed
- Bargmann C. I., Hartwieg E., Horvitz H. R. Odorant selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials