MetaGene: prokaryotic gene finding from environmental genome shotgun sequences - PubMed (original) (raw)
MetaGene: prokaryotic gene finding from environmental genome shotgun sequences
Hideki Noguchi et al. Nucleic Acids Res. 2006.
Abstract
Exhaustive gene identification is a fundamental goal in all metagenomics projects. However, most metagenomic sequences are unassembled anonymous fragments, and conventional gene-finding methods cannot be applied. We have developed a prokaryotic gene-finding program, MetaGene, which utilizes di-codon frequencies estimated by the GC content of a given sequence with other various measures. MetaGene can predict a whole range of prokaryotic genes based on the anonymous genomic sequences of a few hundred bases, with a sensitivity of 95% and a specificity of 90% for artificial shotgun sequences (700 bp fragments from 12 species). MetaGene has two sets of codon frequency interpolations, one for bacteria and one for archaea, and automatically selects the proper set for a given sequence using the domain classification method we propose. The domain classification works properly, correctly assigning domain information to more than 90% of the artificial shotgun sequences. Applied to the Sargasso Sea dataset, MetaGene predicted almost all of the annotated genes and a notable number of novel genes. MetaGene can be applied to wide variety of metagenomic projects and expands the utility of metagenomics.
Figures
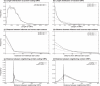
Figure 1
Length distributions of (a) annotated ORFs and (b) random ORFs. ORFs are classified by the GC% of their genomes, and then the length distributions of each class are calculated. Distributions of distances between (c) leftmost start codons and annotated start codons and (d) leftmost start codons and incorrect start codons. Zero means the leftmost start codons are used as the start codons of the ORFs. (e) Orientation-dependent distributions of distances between neighboring ORFs. (f) The background distributions. Negative values mean overlapping of ORFs.
Figure 2
Distributions of log-odds scores of ORFs calculated with (a) archaeal models and (b) bacterial models. Score distributions of the annotated genes and false positives for Archaeoglobus fulgidus and Escherichia coli are indicated.
Figure 3
Sensitivity and specificity of MetaGene for the sets of fixed-length artificial shotgun sequences. The average values for 12 species are indicated.
Figure 4
Effect of sequence errors on gene-finding. Nucleotides of the artificial sequences (700 bp) were changed according to position-specific error rates derived from actual data. The percentages are plotted against the averages of the position-specific error rates.
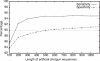
Figure 5
Length distributions of ORFs predicted by various methods. The theoretical distribution was calculated by using gene densities and the length distributions of complete ORFs.
References
- Rappe M., Giovannoni S. The uncultured microbial majority. Annu. Rev. Microbiol. 2003;57:369–394. - PubMed
- Tyson G.W., Chapman J., Hugenholtz P., Allen E.E., Ram R.J., Richardson P.M., Solovyev V.V., Rubin E.M., Rokhsar D.S., Banfield J.F. Community structure and metabolism through reconsruction of microbial genomes from the environment. Nature. 2004;428:37–43. - PubMed
- Venter J.C., Remington K., Heidelberg J.F., Halpern A.L., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. - PubMed
- Tringle S.G., Mering C.V., Kobayashi A., Salamov A.A., Chen K., Chang H.W., Podar M., Short J.M., Mathur E.J., Detter J.C., et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous