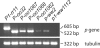Transposition of reversed Ac element ends generates novel chimeric genes in maize - PubMed (original) (raw)
Transposition of reversed Ac element ends generates novel chimeric genes in maize
Jianbo Zhang et al. PLoS Genet. 2006.
Abstract
The maize Activator/Dissociation (Ac/Ds) elements are members of the hAT (hobo, Ac, and Tam3) superfamily of type II (DNA) transposons that transpose through a "cut-and-paste" mechanism. Previously, we reported that a pair of Ac ends in reversed orientation is capable of undergoing alternative transposition reactions that can generate large-scale chromosomal rearrangements, including deletions and inversions. We show here that rearrangements induced by reversed Ac ends transposition can join the coding and regulatory sequences of two linked paralogous genes to generate a series of chimeric genes, some of which are functional. To our knowledge, this is the first report demonstrating that alternative transposition reactions can recombine gene segments, leading to the creation of new genes.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
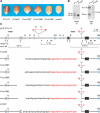
Figure 1. Phenotypes and Gene Structures of P-oo Alleles
(A) The kernel pericarp pigmentation phenotypes specified by the indicated alleles. (B) Genomic Southern blot. Genomic DNA from plants homozygous for the indicated alleles was cut with KpnI and HindIII, and hybridized with probes 15 or 8B from the p1 gene. Lanes marked P-oo32 contain approximately twice as much DNA as lanes marked P1-rr11; this DNA overloading enables the detection of the 7.6-kb band in the KpnI 8B blot, but also results in the intense 6.5-kb band in the HindIII 15 blot. (C) Restriction map. The solid and gray boxes are exons 1, 2, and 3 (left to right) of p1 and p2, respectively. Red triangles indicate Ac or fAc insertions, and the open and solid arrowheads indicate the 3′ and 5′ ends, respectively, of Ac/fAc. Sequences hybridizing with Southern blot probes are indicated by the solid bars above (probe 8B) and below (probe 15) the map. The short horizontal arrows indicate the orientations and approximate position of PCR primers. Primers are identified by numbers below the arrows. The sequence of the junction of each fusion allele is shown here; the black letters indicate p2 sequence, while the red letters indicate fAc sequence. K, KpnI; H, HindIII. Lines below the map indicate the restriction fragments produced by digestion with KpnI or HindIII and hybridizing with the indicated probe; asterisks indicate HindIII restriction sites located within Ac or fAc sequences.
Figure 2. Deletions by Reversed Ac Ends Transposition Generate Chimerical Genes
The solid circle indicates the centromere, the short vertical line indicates the target site, and the other symbols have the same meaning as those in Figure 1. (For animated version, see Video S1). (A) Ac transposase (blue oval) binds to the 5′ end of Ac and 3′ end of fAc. (B) As in ordinary transposition, the Ac 5′ end and the fAc 3′ end are excised by transposase cleavage, and the sequences flanking the Ac/fAc ends join together to form a ~13-kb circle. The X mark at the junction indicates the transposon footprint. (C) The excised transposon ends insert into a site in intron 2 of p2. The Ac 5′ end joins to the distal side of the insertion site to form a circle, and the fAc 3′ end joins to the proximal side of the insertion site to generate a chimeric gene containing exon 1 and exon 2 of p2 and exon 3 of p1. This study reports the isolation of the progenitor (A) and deletion products (C). Note that the hypothetical structures shown in (B) are transient in nature and would not be amenable to physical isolation.
Figure 3. RT-PCR Analysis of P-oo Transcripts
RNA was extracted from kernel pericarp (20 DAP), reverse transcribed, and PCR-amplified using primers complementary to both p1 and p2 transcripts. The progenitor allele (P1-rr11) shows amplification of a 605-bp band from p1. The p-ww2 and P-oo alleles show amplification of a 522-bp band characteristic of the 5′ region of the p2 gene. The p1-ww1112 allele has a deletion of p1; the native p2 gene is intact in this allele, but is not expressed in kernel pericarp.
Similar articles
- Alternative Transposition Generates New Chimeric Genes and Segmental Duplications at the Maize p1 Locus.
Wang D, Yu C, Zuo T, Zhang J, Weber DF, Peterson T. Wang D, et al. Genetics. 2015 Nov;201(3):925-35. doi: 10.1534/genetics.115.178210. Epub 2015 Oct 4. Genetics. 2015. PMID: 26434719 Free PMC article. - Transposition of maize Ac/Ds transposable elements in the yeast Saccharomyces cerevisiae.
Weil CF, Kunze R. Weil CF, et al. Nat Genet. 2000 Oct;26(2):187-90. doi: 10.1038/82827. Nat Genet. 2000. PMID: 11017074 - Transposition of reversed Ac element ends generates chromosome rearrangements in maize.
Zhang J, Peterson T. Zhang J, et al. Genetics. 2004 Aug;167(4):1929-37. doi: 10.1534/genetics.103.026229. Genetics. 2004. PMID: 15342530 Free PMC article. - Molecular biology of maize Ac/Ds elements: an overview.
Lazarow K, Doll ML, Kunze R. Lazarow K, et al. Methods Mol Biol. 2013;1057:59-82. doi: 10.1007/978-1-62703-568-2_5. Methods Mol Biol. 2013. PMID: 23918421 Review. - How maize transposable elements escape negative selection.
Gierl A. Gierl A. Trends Genet. 1990 May;6(5):155-8. doi: 10.1016/0168-9525(90)90150-5. Trends Genet. 1990. PMID: 2164267 Review.
Cited by
- Identification, characterization and distribution of transposable elements in the flax (Linum usitatissimum L.) genome.
González LG, Deyholos MK. González LG, et al. BMC Genomics. 2012 Nov 21;13:644. doi: 10.1186/1471-2164-13-644. BMC Genomics. 2012. PMID: 23171245 Free PMC article. - Small RNA-Mediated De Novo Silencing of Ac/Ds Transposons Is Initiated by Alternative Transposition in Maize.
Wang D, Zhang J, Zuo T, Zhao M, Lisch D, Peterson T. Wang D, et al. Genetics. 2020 Jun;215(2):393-406. doi: 10.1534/genetics.120.303264. Epub 2020 Apr 21. Genetics. 2020. PMID: 32317287 Free PMC article. - Complex chromosomal rearrangements induced by transposons in maize.
Sharma SP, Peterson T. Sharma SP, et al. Genetics. 2023 Feb 9;223(2):iyac124. doi: 10.1093/genetics/iyac124. Genetics. 2023. PMID: 36111993 Free PMC article. - Transposable elements: powerful contributors to angiosperm evolution and diversity.
Oliver KR, McComb JA, Greene WK. Oliver KR, et al. Genome Biol Evol. 2013;5(10):1886-901. doi: 10.1093/gbe/evt141. Genome Biol Evol. 2013. PMID: 24065734 Free PMC article. - Excision and reinsertion of Ac macrotransposons in maize.
Wang D, Yu C, Zhang J, Peterson T. Wang D, et al. Genetics. 2022 Jul 30;221(4):iyac067. doi: 10.1093/genetics/iyac067. Genetics. 2022. PMID: 35471241 Free PMC article.
References
- Kunze R, Weil CF. The hAT and CACTA superfamilies of plant transposons. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington (D. C.): ASM Press; 2002. pp. 565–610.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources