Comparative genomics of the lactic acid bacteria - PubMed (original) (raw)
. 2006 Oct 17;103(42):15611-6.
doi: 10.1073/pnas.0607117103. Epub 2006 Oct 9.
A Slesarev, Y Wolf, A Sorokin, B Mirkin, E Koonin, A Pavlov, N Pavlova, V Karamychev, N Polouchine, V Shakhova, I Grigoriev, Y Lou, D Rohksar, S Lucas, K Huang, D M Goodstein, T Hawkins, V Plengvidhya, D Welker, J Hughes, Y Goh, A Benson, K Baldwin, J-H Lee, I Díaz-Muñiz, B Dosti, V Smeianov, W Wechter, R Barabote, G Lorca, E Altermann, R Barrangou, B Ganesan, Y Xie, H Rawsthorne, D Tamir, C Parker, F Breidt, J Broadbent, R Hutkins, D O'Sullivan, J Steele, G Unlu, M Saier, T Klaenhammer, P Richardson, S Kozyavkin, B Weimer, D Mills
Affiliations
- PMID: 17030793
- PMCID: PMC1622870
- DOI: 10.1073/pnas.0607117103
Comparative genomics of the lactic acid bacteria
K Makarova et al. Proc Natl Acad Sci U S A. 2006.
Abstract
Lactic acid-producing bacteria are associated with various plant and animal niches and play a key role in the production of fermented foods and beverages. We report nine genome sequences representing the phylogenetic and functional diversity of these bacteria. The small genomes of lactic acid bacteria encode a broad repertoire of transporters for efficient carbon and nitrogen acquisition from the nutritionally rich environments they inhabit and reflect a limited range of biosynthetic capabilities that indicate both prototrophic and auxotrophic strains. Phylogenetic analyses, comparison of gene content across the group, and reconstruction of ancestral gene sets indicate a combination of extensive gene loss and key gene acquisitions via horizontal gene transfer during the coevolution of lactic acid bacteria with their habitats.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
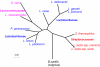
Fig. 1.
Phylogenetic trees of Lactobacillales constructed on the basis of concatenated alignments of ribosomal proteins. All branches are supported at >75% bootstrap values. Species are colored according to the current taxonomy: Lactobacillaceae, blue; Leuconostocaceae, magenta; Streptococcaceae, red.
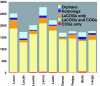
Fig. 2.
Conserved and unique genes in the genomes of Lactobacillales. “Homolog” indicates genes with detectable homologs in organisms other than those analyzed here but could not be included in any of the COG sets. “Orphans” are predicted genes without detectable homologs. Lacga, Lb. gasseri; Lacbr, Lb. brevis; Pedpe, P. pentosaceus; Laccr, Lc. lactis ssp. cremoris; Strth, S. thermophilus; Oenoe, O. oeni; Leume, Le. mesenteroides; Lacca, Lb. casei; Lacde, Lb. delbrueckii; Lacla, Lc. lactis; Lacpl, Lb. plantarum; Lacjo, Lb. johnsonii. The vertical axis shows the number of genes per genome.
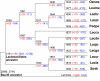
Fig. 3.
Reconstruction of gene content evolution in Lactobacillales. The tree topology is as in Fig. 1, rooted by using Baccillus subtilis as the outgroup. For each species and each internal node of tree, the inferred number of LaCOGs present, and the numbers of LaCOGs lost (blue) and gained (red) along the branch leading to the given node (species) are indicated. Abbreviations are as in Fig. 2.
Similar articles
- Evolutionary genomics of lactic acid bacteria.
Makarova KS, Koonin EV. Makarova KS, et al. J Bacteriol. 2007 Feb;189(4):1199-208. doi: 10.1128/JB.01351-06. Epub 2006 Nov 3. J Bacteriol. 2007. PMID: 17085562 Free PMC article. Review. No abstract available. - Genetics of lactic acid bacteria with special reference to lactococci.
Rodríguez A, Vidal DR. Rodríguez A, et al. Microbiologia. 1990 Dec;6(2):51-64. Microbiologia. 1990. PMID: 2095168 Review. - Advances in the genomics and metabolomics of dairy lactobacilli: A review.
Stefanovic E, Fitzgerald G, McAuliffe O. Stefanovic E, et al. Food Microbiol. 2017 Feb;61:33-49. doi: 10.1016/j.fm.2016.08.009. Epub 2016 Aug 31. Food Microbiol. 2017. PMID: 27697167 Review. - From physiology to systems metabolic engineering for the production of biochemicals by lactic acid bacteria.
Gaspar P, Carvalho AL, Vinga S, Santos H, Neves AR. Gaspar P, et al. Biotechnol Adv. 2013 Nov;31(6):764-88. doi: 10.1016/j.biotechadv.2013.03.011. Epub 2013 Apr 6. Biotechnol Adv. 2013. PMID: 23567148 Review. - Lactic acid bacteria of foods and their current taxonomy.
Stiles ME, Holzapfel WH. Stiles ME, et al. Int J Food Microbiol. 1997 Apr 29;36(1):1-29. doi: 10.1016/s0168-1605(96)01233-0. Int J Food Microbiol. 1997. PMID: 9168311 Review.
Cited by
- Metabolome and Metagenome Integration Unveiled Synthesis Pathways of Novel Antioxidant Peptides in Fermented Lignocellulosic Biomass of Palm Kernel Meal.
Qamar H, He R, Li Y, Song M, Deng D, Cui Y, Yu M, Ma X. Qamar H, et al. Antioxidants (Basel). 2024 Oct 17;13(10):1253. doi: 10.3390/antiox13101253. Antioxidants (Basel). 2024. PMID: 39456506 Free PMC article. - Use of Rgg quorum-sensing machinery to create an innovative recombinant protein expression system in Streptococcus thermophilus.
Gardan R, Honvo-Houeto E, Mézange C, Maillot NJ, Balvay A, Rabot S, Bermúdez-Humarán LG, Langella P, Monnet V, Juillard V. Gardan R, et al. Microbiology (Reading). 2024 Sep;170(9):001487. doi: 10.1099/mic.0.001487. Microbiology (Reading). 2024. PMID: 39302176 Free PMC article. - Functional and practical insights into three lactococcal antiphage systems.
Grafakou A, Mosterd C, de Waal PP, van Rijswijck IMH, van Peij NNME, Mahony J, van Sinderen D. Grafakou A, et al. Appl Environ Microbiol. 2024 Sep 18;90(9):e0112024. doi: 10.1128/aem.01120-24. Epub 2024 Aug 13. Appl Environ Microbiol. 2024. PMID: 39136492 - Discovery of antiphage systems in the lactococcal plasmidome.
Grafakou A, Mosterd C, Beck MH, Kelleher P, McDonnell B, de Waal PP, van Rijswijck IMH, van Peij NNME, Cambillau C, Mahony J, van Sinderen D. Grafakou A, et al. Nucleic Acids Res. 2024 Sep 9;52(16):9760-9776. doi: 10.1093/nar/gkae671. Nucleic Acids Res. 2024. PMID: 39119896 Free PMC article. - Liquorilactobacillus: A Context of the Evolutionary History and Metabolic Adaptation of a Bacterial Genus from Fermentation Liquid Environments.
da Silva Santos D, Freitas NSA, de Morais MA Jr, Mendonça AA. da Silva Santos D, et al. J Mol Evol. 2024 Aug;92(4):467-487. doi: 10.1007/s00239-024-10189-6. Epub 2024 Jul 17. J Mol Evol. 2024. PMID: 39017924
References
- Miller N, Wetterstrom W. In: The Cambridge World History of Food. Kiple K, Ornelas K, editors. Vol 2. Cambridge, UK: Cambridge Univ Press; 2000. pp. 1123–1139.
- Wood B. Microbiology of Fermented Foods. London: Blackie; 1998.
- Kandler O. Antonie van Leeuwenhoek. 1983;49:209–224. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases