Antibody recognition and neutralization determinants on domains I and II of West Nile Virus envelope protein - PubMed (original) (raw)
Comparative Study
. 2006 Dec;80(24):12149-59.
doi: 10.1128/JVI.01732-06. Epub 2006 Oct 11.
Grant E Nybakken, Michael Engle, Qing Xu, Christopher A Nelson, Soila Sukupolvi-Petty, Anantha Marri, Bat-El Lachmi, Udy Olshevsky, Daved H Fremont, Theodore C Pierson, Michael S Diamond
Affiliations
- PMID: 17035317
- PMCID: PMC1676294
- DOI: 10.1128/JVI.01732-06
Comparative Study
Antibody recognition and neutralization determinants on domains I and II of West Nile Virus envelope protein
Theodore Oliphant et al. J Virol. 2006 Dec.
Abstract
Previous studies have demonstrated that monoclonal antibodies (MAbs) against an epitope on the lateral surface of domain III (DIII) of the West Nile virus (WNV) envelope (E) strongly protect against infection in animals. Herein, we observed significantly less efficient neutralization by 89 MAbs that recognized domain I (DI) or II (DII) of WNV E protein. Moreover, in cells expressing Fc gamma receptors, many of the DI- and DII-specific MAbs enhanced infection over a broad range of concentrations. Using yeast surface display of E protein variants, we identified 25 E protein residues to be critical for recognition by DI- or DII-specific neutralizing MAbs. These residues cluster into six novel and one previously characterized epitope located on the lateral ridge of DI, the linker region between DI and DIII, the hinge interface between DI and DII, and the lateral ridge, central interface, dimer interface, and fusion loop of DII. Approximately 45% of DI-DII-specific MAbs showed reduced binding with mutations in the highly conserved fusion loop in DII: 85% of these (34 of 40) cross-reacted with the distantly related dengue virus (DENV). In contrast, MAbs that bound the other neutralizing epitopes in DI and DII showed no apparent cross-reactivity with DENV E protein. Surprisingly, several of the neutralizing epitopes were located in solvent-inaccessible positions in the context of the available pseudoatomic model of WNV. Nonetheless, DI and DII MAbs protect against WNV infection in mice, albeit with lower efficiency than DIII-specific neutralizing MAbs.
Figures
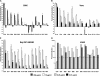
FIG. 1.
In vitro neutralization and enhancement activity of DI- or DII-specific MAbs. (A) Neutralization of WNV using a PRNT assay on BHK cells. (B) Neutralization of RVPs on Vero cells. (C) Neutralization of RVPs on Raji DC-SIGNR cells. The data are expressed as percentages of the no-MAb control. The data shown are the means from at least three independent experiments. Error bars indicate the standard errors of the means, and statistical significance was determined using an unpaired, two-tailed t test compared to the no-MAb control (*, P ≤ 0.05; **, P ≤ 0.01). (D) Enhancement of RVP infection on K562 cells. The data shown are the enhancement over baseline infection without MAb. All MAbs were studied in a single experiment to allow for comparisons of the power of enhancement. Values of less than 100 indicate neutralization, whereas values greater than 100 indicate enhancement.
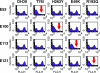
FIG. 2.
Flow cytometry patterns of loss-of-function DI- or DII-specific MAb variants selected by yeast surface display. Representative histograms are shown for MAbs E53, E100, E113, and E121. Red arrows indicate mutations that result in loss of MAb binding. The data shown are representative of three independent experiments. FL4-H, log fluorescence intensity on the FL4 (660=nm) channel.
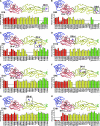
FIG. 3.
Epitope mapping of DI- and DII-specific neutralizing MAbs. Binding of (A) E18, (B) E53, (C) 7H7, (D) E113, (E) E121, (F) E48, (G) E100, and (H) E101 to mutants expressed on the yeast surface. The binding of each MAb to the mutants was measured by flow cytometry, and total fluorescence was normalized to yeast expressing wild-type DI-DII. The data shown are the means from three independent experiments. Error bars indicate the standard errors of the means. The colors red, yellow, blue, and green indicate domains I, II, and III and the fusion loop, respectively. Mutations that resulted in ≥50% reduction of MAb binding were mapped (shown in magenta and boxed) onto the WNV E protein crystal structure (Protein Data Bank accession code 2HG0). For MAbs E53 and 7H7, residues that compose the primary binding site within DII are boxed and secondary sites in DI are circled. Epitopes are labeled using the same nomenclature defined in Table 1.
FIG. 4.
Epitope expression on the WNV virion. Yeast display epitope residues (magenta) for (A) E16, (B) E18, (C) E53, (D) 7H7, (E) E113, and (F) E121 were mapped onto the pseudoatomic model of the mature WNV virion. For E16, the blue indicates additional contact residues as determined by X-ray crystallography. Virions are depicted as 2.0-Å-radius Cα atoms and are colored according to their symmetry axes, twofold (cyan), threefold (green), and fivefold (yellow). Epitopes are boxed on one E protein in each symmetry axis. Secondary binding sites in DI for E53 and 7H7 are circled in each symmetry axis.
Similar articles
- Human monoclonal antibodies against West Nile virus induced by natural infection neutralize at a postattachment step.
Vogt MR, Moesker B, Goudsmit J, Jongeneelen M, Austin SK, Oliphant T, Nelson S, Pierson TC, Wilschut J, Throsby M, Diamond MS. Vogt MR, et al. J Virol. 2009 Jul;83(13):6494-507. doi: 10.1128/JVI.00286-09. Epub 2009 Apr 22. J Virol. 2009. PMID: 19386704 Free PMC article. - Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes.
Sukupolvi-Petty S, Austin SK, Purtha WE, Oliphant T, Nybakken GE, Schlesinger JJ, Roehrig JT, Gromowski GD, Barrett AD, Fremont DH, Diamond MS. Sukupolvi-Petty S, et al. J Virol. 2007 Dec;81(23):12816-26. doi: 10.1128/JVI.00432-07. Epub 2007 Sep 19. J Virol. 2007. PMID: 17881453 Free PMC article. - Induction of epitope-specific neutralizing antibodies against West Nile virus.
Oliphant T, Nybakken GE, Austin SK, Xu Q, Bramson J, Loeb M, Throsby M, Fremont DH, Pierson TC, Diamond MS. Oliphant T, et al. J Virol. 2007 Nov;81(21):11828-39. doi: 10.1128/JVI.00643-07. Epub 2007 Aug 22. J Virol. 2007. PMID: 17715236 Free PMC article. - The molecular basis of antibody-mediated neutralization of West Nile virus.
Oliphant T, Diamond MS. Oliphant T, et al. Expert Opin Biol Ther. 2007 Jun;7(6):885-92. doi: 10.1517/14712598.7.6.885. Expert Opin Biol Ther. 2007. PMID: 17555373 Review. - The structural immunology of antibody protection against West Nile virus.
Diamond MS, Pierson TC, Fremont DH. Diamond MS, et al. Immunol Rev. 2008 Oct;225:212-25. doi: 10.1111/j.1600-065X.2008.00676.x. Immunol Rev. 2008. PMID: 18837784 Free PMC article. Review.
Cited by
- Adaptive Immunity to Dengue Virus: Slippery Slope or Solid Ground for Rational Vaccine Design?
Wilken L, Rimmelzwaan GF. Wilken L, et al. Pathogens. 2020 Jun 15;9(6):470. doi: 10.3390/pathogens9060470. Pathogens. 2020. PMID: 32549226 Free PMC article. Review. - Persistence of circulating memory B cell clones with potential for dengue virus disease enhancement for decades following infection.
Smith SA, Zhou Y, Olivarez NP, Broadwater AH, de Silva AM, Crowe JE Jr. Smith SA, et al. J Virol. 2012 Mar;86(5):2665-75. doi: 10.1128/JVI.06335-11. Epub 2011 Dec 14. J Virol. 2012. PMID: 22171265 Free PMC article. - The mechanistic basis of protection by non-neutralizing anti-alphavirus antibodies.
Earnest JT, Holmes AC, Basore K, Mack M, Fremont DH, Diamond MS. Earnest JT, et al. Cell Rep. 2021 Apr 6;35(1):108962. doi: 10.1016/j.celrep.2021.108962. Cell Rep. 2021. PMID: 33826892 Free PMC article. - The infectivity of prM-containing partially mature West Nile virus does not require the activity of cellular furin-like proteases.
Mukherjee S, Lin TY, Dowd KA, Manhart CJ, Pierson TC. Mukherjee S, et al. J Virol. 2011 Nov;85(22):12067-72. doi: 10.1128/JVI.05559-11. Epub 2011 Aug 31. J Virol. 2011. PMID: 21880759 Free PMC article. - Human monoclonal antibodies against West Nile virus induced by natural infection neutralize at a postattachment step.
Vogt MR, Moesker B, Goudsmit J, Jongeneelen M, Austin SK, Oliphant T, Nelson S, Pierson TC, Wilschut J, Throsby M, Diamond MS. Vogt MR, et al. J Virol. 2009 Jul;83(13):6494-507. doi: 10.1128/JVI.00286-09. Epub 2009 Apr 22. J Virol. 2009. PMID: 19386704 Free PMC article.
References
- Beasley, D. W., and J. G. Aaskov. 2001. Epitopes on the dengue 1 virus envelope protein recognized by neutralizing IgM monoclonal antibodies. Virology 279:447-458. - PubMed
- Ben-Nathan, D., S. Lustig, G. Tam, S. Robinzon, S. Segal, and B. Rager-Zisman. 2003. Prophylactic and therapeutic efficacy of human intravenous immunoglobulin in treating West Nile virus infection in mice. J Infect. Dis. 188:5-12. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases