Community structure analyses are more sensitive to differences in soil bacterial communities than anonymous diversity indices - PubMed (original) (raw)
Community structure analyses are more sensitive to differences in soil bacterial communities than anonymous diversity indices
Martin Hartmann et al. Appl Environ Microbiol. 2006 Dec.
Abstract
Changes in the diversity and structure of soil microbial communities may offer a key to understanding the impact of environmental factors on soil quality in agriculturally managed systems. Twenty-five years of biodynamic, bio-organic, or conventional management in the DOK long-term experiment in Switzerland significantly altered soil bacterial community structures, as assessed by terminal restriction fragment length polymorphism (T-RFLP) analysis. To evaluate these results, the relation between bacterial diversity and bacterial community structures and their discrimination potential were investigated by sequence and T-RFLP analyses of 1,904 bacterial 16S rRNA gene clones derived from the DOK soils. Standard anonymous diversity indices such as Shannon, Chao1, and ACE or rarefaction analysis did not allow detection of management-dependent influences on the soil bacterial community. Bacterial community structures determined by sequence and T-RFLP analyses of the three gene libraries substantiated changes previously observed by soil bacterial community level T-RFLP profiling. This supported the value of high-throughput monitoring tools such as T-RFLP analysis for assessment of differences in soil microbial communities. The gene library approach also allowed identification of potential management-specific indicator taxa, which were derived from nine different bacterial phyla. These results clearly demonstrate the advantages of community structure analyses over those based on anonymous diversity indices when analyzing complex soil microbial communities.
Figures
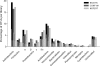
FIG. 1.
Relative abundance of OTUs detected in the three soil bacterial 16S rRNA gene libraries obtained from three farming systems of the DOK field experiment. Numbers of OTUs for each bacterial phylum and gene library are displayed as percentages of the corresponding gene library size, i.e., BIODYN (100% = 600 sequences), CONFYM (100% = 691 sequences), and NOFERT (100% = 613 sequences). Three different OTU definitions, i.e., PSIL97-OTU (filled bars), PSIL90-OTU (small white framed bars), and T-RFseq-OTU (narrow white bars), were applied.
FIG. 2.
Rarefaction analysis of soil bacterial 16S rRNA gene libraries from three agricultural farming systems of the DOK field experiment, displaying number of OTUs detected versus number of sequences analyzed. Aligned sequences were grouped using FastGroup v1.2 software at different OTU definitions, i.e., PSIL97-OTU, PSIL90-OTU, and PSIL80-OTU. In addition, an OTU definition based on unique in silico T-RFseq sizes is displayed. Rarefaction curves are displayed for each OTU definition.
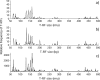
FIG. 3.
T-RFLP profiles of bacterial 16S rRNA genes from DNA of CONFYM in the DOK field experiment. (a) Experimental T-RFLP profile compiled from T-RFexp of each clone in the corresponding gene library yielding T-RFexp between 50 and 500 rmu. (b) In silico T-RFLP profile compiled from sequence-predicted T-RFseq of each clone in the corresponding gene library yielding T-RFseq between 50 and 500 bp (c) Bacterial community level T-RFLP profile from soil DNA extracts in a range from 50 to 500 rmu as obtained in a previous study (21).
FIG. 4.
Differences of bacterial 16S rRNA gene-based community structures between the three farming systems in the DOK experiment as determined by Ward clustering of Euclidean distances. Cluster dendrograms are based on OTUs derived from 16S rRNA gene libraries and from soil bacterial community level T-RFLP profiles. OTUs were defined at a PSIL of 97 with 899 OTUs (a) and at a PSIL of 90 with 272 OTUs (b). (c) Cluster analysis of soil bacterial community level T-RFLP profiles of pooled DNA extracts from four field replicates and based on intensities of 79 scorable T-RF sizes among four field replicates (community T-RFLP) (21).
References
- Bardgett, R. D. 2002. Causes and consequences of biological diversity in soil. Zoology 105:367-374. - PubMed
- Bengtsson, J. 1998. Which species? What kind of diversity? Which ecosystem function? Some problems in studies of relations between biodiversity and ecosystem function. Appl. Soil Ecol. 10:191-199.
- Bohannan, B. J. M., and J. Hughes. 2003. New approaches to analyzing microbial biodiversity data. Curr. Opin. Microbiol. 6:282-287. - PubMed
- Bossio, D. A., K. M. Scow, N. Gunapala, and K. J. Graham. 1998. Determinants of soil microbial communities: effects of agricultural management, season, and soil type on phospholipid fatty acid profiles. Microb. Ecol. 36:1-12. - PubMed
- Brown, J. H., S. K. M. Ernest, J. M. Parody, and J. P. Haskell. 2001. Regulation of diversity: maintenance of species richness in changing environments. Oecologia 126:321-332. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous