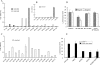Identifying epitopes responsible for neutralizing antibody and DC-SIGN binding on the spike glycoprotein of the severe acute respiratory syndrome coronavirus - PubMed (original) (raw)
Identifying epitopes responsible for neutralizing antibody and DC-SIGN binding on the spike glycoprotein of the severe acute respiratory syndrome coronavirus
Yi-Ping Shih et al. J Virol. 2006 Nov.
Abstract
The severe acute respiratory syndrome-associated coronavirus (SARS-CoV) uses dendritic cell-specific ICAM-3 grabbing nonintegrin (DC-SIGN) to facilitate cell entry via cellular receptor-angiotensin-converting enzyme 2. For this project, we used recombinant baculoviruses expressing different lengths of SARS-CoV spike (S) protein in a capture assay to deduce the minimal DC-SIGN binding region. Our results identified the region location between amino acid (aa) residues 324 to 386 of the S protein. We then generated nine monoclonal antibodies (MAbs) against the S protein to map the DC-SIGN-binding domain using capture assays with pseudotyped viruses and observed that MAb SIa5 significantly blocked S protein-DC-SIGN interaction. An enhancement assay using the HKU39849 SARS-CoV strain and human immature dendritic cells confirmed our observation. Data from a pepscan analysis and M13 phage peptide display library system mapped the reactive MAb SIa5 epitope to aa residues 363 to 368 of the S protein. Results from a capture assay testing three pseudotyped viruses with mutated N-linked glycosylation sites of the S protein indicate that only two pseudotyped viruses (N330Q and N357Q, both of which lost glycosylation sites near the SIa5 epitope) had diminished DC-SIGN-binding capacity. We also noted that MAb SIb4 exerted a neutralizing effect against HKU39849; its reactive epitope was mapped to aa residues 435 to 439 of the S protein. We offer the data to facilitate the development of therapeutic agents and preventive vaccines against SARS-CoV infection.
Figures
FIG. 1.
DC-SIGN binding domain identification in the SARS-CoV S protein using a panel of recombinant baculoviruses. (A) Direct infection assay. A recombinant baculovirus-vAtEpGS763 containing 17 to 763 aa residues of SARS-CoV S protein was used to infect Vero E6, B-THP-1, and B-THP-1/DC-SIGN cells. (B) Capture assay. B-THP-1 or B-THP-1/DC-SIGN cells were incubated with a wild-type virus (vAtE) or a panel of recombinant baculoviruses containing different lengths of SARS-CoV S protein (MOI = 10). After 2 washings, cells were cocultured with Vero E6 cells for 48 h. #, P < 0.05, Student's t test. (C) Specificity of the interaction between vAtEpGS386 and DC-SIGN. Recombinant baculovirus vAtEpGS386 was incubated with B-THP-1/DC-SIGN cells in the presence of mannan (20 μg/ml−1), EGTA (5 mM), or anti-DC-SIGN MAb (10 μg/ml−1). The unbound virus was eliminated by repeated washings prior to coculture with Vero E6 cells. Infections by recombinant baculoviruses were determined by measuring EGFP expression using flow cytometry. (D) Virus binding assay. B-THP-1 or B-THP-1/DC-SIGN cells were incubated with different recombinant baculoviruses for 2 h before subjected to flow cytometry with anti-S antibody.
FIG. 2.
DC-SIGN not only mediates but also enhances pseudotyped virus infection to HepG2 cells. (A) Direct infection assay. HepG2, B-THP-1, and B-THP-1/DC-SIGN were directly infected with pseudotyped virus equivalent to 4 ng p24 antigen. (B) Capture assay. B-THP-1 or B-THP-1/DC-SIGN cells were incubated with pseudotyped virus for 2 h, washed twice, and then cocultured with HepG2 cells. To study the specificity of the interaction, either B-THP-1 or B-THP-1/DC-SIGN cells were preincubated with mannan (20 μg/ml−1), sucrose (20 μg/ml−1), EGTA (5 mM), anti-DC-SIGN MAb (10 μg/ml−1), or anti-L-SIGN MAb (10 μg/ml−1) at 37°C for 1 h before the addition of pseudotyped viruses. *, P < 0.01, Student's t test. Among all the capture assays with B-THP-1/DC-SIGN (black bars), the luciferase counts of the assays preincubated with EGTA, mannan, or anti-DC-SIGN were significantly lower than that of the medium control (P < 0.01). (C) Enhancement assay. B-THP-1 and B-THP-1/DC-SIGN cells were incubated with pseudotyped virus and then cocultured with HepG2 cells without washing. To study the specificity of the enhancement, either B-THP-1 or B-THP-1/DC-SIGN cells were preincubated with mannan (20 μg/ml−1), sucrose (20 μg/ml−1), EGTA (5 mM), anti-L-SIGN MAb (10 μg/ml−1), or anti-DC-SIGN MAb (10 μg/ml−1) before the addition of pseudotyped viruses. The infectivity was determined 48 h after the infection in the HepG2 cells by measuring the luciferase activity. One representative experiment of two is shown. #, P < 0.05, Student's t test.
FIG. 3.
Neutralizing effect of MAb SIb4. (A) A pseudotyped virus was preincubated with each MAb (15 μg/ml) for 1 h at 37°C prior to infection of HepG2 cells. Infectivity was determined 48 h postinfection by luciferase activity measurement. (B) SARS-CoV strain HKU39849 (3.125 TCID50) was incubated with mIgG (a), SIb2 (b), or SIb4 (c) for 1 h at 37°C and then added to Vero E6 cells. Images were taken 96 h postinfection. (C) ORF1b copy numbers in culture supernatant were determined using real-time RT-PCR. Results from one representative experiment of two are shown.
FIG. 4.
MAb SIa5 interrupts SARS-CoV S protein interacted with DC-SIGN in a capture assay. (A) Pseudotyped virus was incubated with each MAb (15 μg/ml) for 1 h at 37°C and then incubated with B-THP-1/DC-SIGN cells for 2 h at 37°C. Cells were washed twice and cocultured with HepG2 cells. The luciferase activity in cell lysates was measured 48 h postinfection. The assay with MAb SIa5 had significantly lower luciferase counts than that with mIgG (P < 0.05). (B) SARS-CoV (50 TCID50) was preincubated with either mIgG (a), SIa10 (b), or SIa5 (c) for 1 h at 37°C, then incubated with B-THP-1/DC-SIGN cells for 2 h at 37°C, washed two times, and added to Vero E6 cells. Images were taken 48 h postinfection. Arrow, B-THP-1/DC-SIGN cells; triangle, Vero E6 cells. (C) Infectivity was determined by measuring the copy numbers of ORF1b mRNA in culture supernatant. Results from one representative experiment of two are shown.
FIG. 5.
MAb SIa5 interrupts SARS-CoV S protein interacted with DC-SIGN in an enhancement assay. (A) SARS-CoV (1.625 TCID50) was mixed with no antibody (b), mIgG (c), or SIa5 (5 μg/ml) (d) for 1 h before being added to immature DCs (iDCs) and cultured for 2 h. Cells were not washed prior to being added to Vero E6 cells. Alternative (a) shows Vero E6 cells directly infected with the SARS-CoV. Images were taken 96 h postinfection. Arrow, B-THP-1/DC-SIGN cells; triangle, Vero E6 cells. (B) Infectivity was determined by measuring the copy numbers of ORF1b mRNA in cultural supernatant. Results from one representative experiment of two are shown.
FIG. 6.
Results of peptide scan for MAbs SIb4 and SIa5. (A) EIAs with 14 SPs, each 50 aa in length, were employed in the study. OD, optical density. (B) EIAs with 8 SPs (each 15 aa in length) were used for MAb SIb4 epitope mapping. (C) EIAs with 16 SPs (each 15 aa in length) were used for MAb SIa5 epitope mapping. (D) Inhibitory effect of SP-434-448 on pseudotyped virus infectivity. Prior to pseudotyped viral infection, HepG2 cells were incubated with SP-434-448, SP-175-205, or OC43-SP-186-200 for 1 h. Infectivity was determined 2 days postinfection by measuring luciferase activity. Results from one representative experiment of two are shown. #, the difference of the luciferase counts between these two reactions was statistically significant (P < 0.05). (E) A lack of glycosylation impaired the ability of the S protein to bind to DC-SIGN. Either B-THP-1 or B-THP-1/DC-SIGN cells were incubated with different pseudotyped viruses for 2 h at 37°C. Cells were washed twice and cocultured with HepG2 cells. Infectivity was determined after 48 h by measuring luciferase activity.
FIG. 7.
Different S protein binding domains for ACE2 and DC-SIGN. Schematic diagram of ACE2 (purple), the carbohydrate recognition domain of DC-SIGN (blue), and binding sites in the receptor binding domain (green) of the SARS-CoV spike protein. Red, MAb SIb4-recognized epitope; orange, MAb SIa5-recognized epitope; yellow, N-glycosylation sites N330 and N357 (modified from reference 29).
Similar articles
- Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants.
ter Meulen J, van den Brink EN, Poon LL, Marissen WE, Leung CS, Cox F, Cheung CY, Bakker AQ, Bogaards JA, van Deventer E, Preiser W, Doerr HW, Chow VT, de Kruif J, Peiris JS, Goudsmit J. ter Meulen J, et al. PLoS Med. 2006 Jul;3(7):e237. doi: 10.1371/journal.pmed.0030237. PLoS Med. 2006. PMID: 16796401 Free PMC article. - Vaccine design for severe acute respiratory syndrome coronavirus.
He Y, Jiang S. He Y, et al. Viral Immunol. 2005;18(2):327-32. doi: 10.1089/vim.2005.18.327. Viral Immunol. 2005. PMID: 16035944 Review. - SARS vaccine development.
Jiang S, He Y, Liu S. Jiang S, et al. Emerg Infect Dis. 2005 Jul;11(7):1016-20. doi: 10.3201/1107.050219. Emerg Infect Dis. 2005. PMID: 16022774 Free PMC article. Review.
Cited by
- Mass Spectrometry Analysis of SARS-CoV-2 Nucleocapsid Protein Reveals Camouflaging Glycans and Unique Post-Translational Modifications.
Sun Z, Zheng X, Ji F, Zhou M, Su X, Ren K, Li L. Sun Z, et al. Infect Microbes Dis. 2021 Aug 3;3(3):149-157. doi: 10.1097/IM9.0000000000000071. eCollection 2021 Sep. Infect Microbes Dis. 2021. PMID: 38630108 Free PMC article. - Sepsis - it is all about the platelets.
Cox D. Cox D. Front Immunol. 2023 Jun 7;14:1210219. doi: 10.3389/fimmu.2023.1210219. eCollection 2023. Front Immunol. 2023. PMID: 37350961 Free PMC article. Review. - Evolutionary Signals in Coronaviral Structural Proteins Suggest Possible Complex Mechanisms of Post-Translational Regulation in SARS-CoV-2 Virus.
Garza-Domínguez R, Torres-Quiroz F. Garza-Domínguez R, et al. Viruses. 2022 Nov 8;14(11):2469. doi: 10.3390/v14112469. Viruses. 2022. PMID: 36366566 Free PMC article. - SARS-CoV-2 S glycoprotein binding to multiple host receptors enables cell entry and infection.
Trbojević-Akmačić I, Petrović T, Lauc G. Trbojević-Akmačić I, et al. Glycoconj J. 2021 Oct;38(5):611-623. doi: 10.1007/s10719-021-10021-z. Epub 2021 Sep 20. Glycoconj J. 2021. PMID: 34542788 Free PMC article. Review. - Targeting SARS-CoV-2-Platelet Interactions in COVID-19 and Vaccine-Related Thrombosis.
Cox D. Cox D. Front Pharmacol. 2021 Jul 5;12:708665. doi: 10.3389/fphar.2021.708665. eCollection 2021. Front Pharmacol. 2021. PMID: 34290613 Free PMC article. Review.
References
- Aden, D. P., A. Fogel, S. Plotkin, I. Damjanov, and B. B. Knowles. 1979. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature 282:615-616. - PubMed
- Bashirova, A. A., T. B. H. Geijtenbeek, G. C. F. van Duijnhoven, S. J. van Vliet, J. B. G. Eilering, M. P. Martin, L. Wu, T. D. Martin, N. Viebig, P. A. Knolle, V. N. KewalRamani, Y. van Kooyk, and M. Carrington. 2001. A dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN)-related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV-1 infection. J. Exp. Med. 193:671-678. - PMC - PubMed
- Chan, V. S. F., K. Y. K. Chan, Y. Chen, L. L. M. Poon, A. N. Y. Cheung, B. Zheng, K. H. Chan, W. Mak, H. Y. S. Ngan, X. Xu, G. Screaton, P. K. H. Tam, J. M. Austyn, L. C. Chan, S. P. Yip, M. Peiris, U. S. Khoo, and C. L. Lin. 2006. Homozygous L-SIGN (CLEC4M) plays a protective role in SARS coronavirus infection. Nat. Genet. 38:38-46. - PMC - PubMed
- Chang, Y. J., C. Y. Y. Liu, B. L. Chiang, Y. C. Chao, and C. C. Chen. 2004. Induction of IL-8 release in lung cells via activator protein-1 by recombinant baculovirus displaying severe acute respiratory syndrome-coronavirus spike proteins: identification of two functional regions. J. Immunol. 173:7602-7614. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous