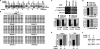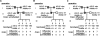Deficiency of Rbbp1/Arid4a and Rbbp1l1/Arid4b alters epigenetic modifications and suppresses an imprinting defect in the PWS/AS domain - PubMed (original) (raw)
Deficiency of Rbbp1/Arid4a and Rbbp1l1/Arid4b alters epigenetic modifications and suppresses an imprinting defect in the PWS/AS domain
Mei-Yi Wu et al. Genes Dev. 2006.
Abstract
Prader-Willi syndrome (PWS) and Angelman syndrome (AS) are caused by deficiency of imprinted gene expression from paternal or maternal chromosome 15q11-q13, respectively. Genomic imprinting of the PWS/AS domain is regulated through a bipartite cis-acting imprinting center (PWS-IC/AS-IC) within and upstream of the SNRPN promoter. Here, we show that two Rb-binding protein-related genes, Rbbp1/Arid4a and Rbbp1l1/Arid4b, are involved in the regulation of imprinting of the IC. We recovered these two genes from gene trap mutagenesis selecting for altered expression of an Snrpn-EGFP fusion gene strategy. RBBP1/ARID4A is an Rb-binding protein. RBBP1/ARID4A interacts with RBBP1L1/ARID4B and with the Snrpn promoter, implying that both are part of a protein complex. To further elucidate their roles on regulation of imprinting, we deleted the Rbbp1/Arid4a and Rbbp1l1/Arid4b genes in mice. Combined homozygous deficiency for Rbbp1/Arid4a and heterozygous deficiency for Rbbp1l1/Arid4b altered epigenetic modifications at the PWS-IC with reduced trimethylation of histone H4K20 and H3K9 and reduced DNA methylation, changing the maternal allele toward a more paternal epigenotype. Importantly, mutations of Rbbp1/Arid4a, Rbbp1l1/Arid4b, or Rb suppressed an AS imprinting defect caused by a mutation at the AS-IC. These data identify Rbbp1/Arid4a and Rbbp1l1/Arid4b as new members of epigenetic complexes regulating genomic imprinting at the PWS/AS domain.
Figures
FIGURE 1.
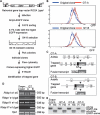
Generation of a _Snrpn_-EGFP fusion gene in ES cells. (A) Targeting strategy for constructing the _Snrpn_-EGFP fusion gene. The EGFP cassette and the neomycin resistance gene (neo) were introduced into exon 3 of Snrpn by homologous recombination. The neo gene, flanked by two loxP sites (triangles), was subsequently removed by the Cre recombinase. (TK) Herpes simplex viral thymidine kinase cassette; (H) HindIII; (P) PstI; (X) XbaI; (Bs) BssHII; (E) EcoRI. (B) Southern blotting analysis of wild-type (+/+) cells, the clone targeted with the EGFP-neo construct (+/EGFP-neo), and the latter cells following Cre-mediated deletion of the neo gene (+/EGFP). (Left) DNA was digested with PstI, and hybridized with the 3′-flanking probe shown in A. (Wt) The 9.0-kb fragment from the wild-type allele; (mt) the 4.3-kb fragment from the mutant EGFP-neo allele. (Right) DNA was digested with EcoRI and hybridized with an EGFP probe. The fragment sizes for EGFP-neo and EGFP alleles are 2.7 kb and 1.0 kb, respectively. (C) Flow cytometric analysis for the comparison of EGFP expression in wild-type ES cells (green), the EGFP fusion gene with neo present (red), and the EGFP clone with neo deleted (blue). The scale is cell number on the left and relative fluorescence below. (D) Methylation analysis of the Snrpn CpG island in the _Snrpn_-EGFP clone. Genomic DNA was digested with HindIII (H) alone or in combination with SacII (HS) for Southern blot analysis. (Wt-me) The 14-kb methylated DNA; (wt-unme) the 10.2-kb unmethylated DNA from the wild-type Snrpn allele; (EGFP-me) the 13.2-kb methylated DNA; (EGFP-unme) the 9.4-kb unmethylated DNA from the _Snrpn_-EGFP fusion allele.
FIGURE 2.
Gene trap mutagenesis using the _Snrpn_-EGFP fusion clone. (A) Scheme for screening mutagenized cells. A retroviral vector, ROSA_β_geo*, was used to infect the _Snrpn_-EGFP clone. The infected cells were sorted by FACS, and the 0.1% of cells with the highest EGFP expression were plated onto the feeder cell plates and selected with G418. The G418-resistant colonies were picked, and the level of EGFP expression for individual clones was confirmed by flow cytometric analysis. Then, identification of the trapped gene was achieved by 5′-RACE. (SA) Splice acceptor site; (_β_geo) a fusion reporter gene including _β_-galactosidase and neomycin resistance activities. (B) Flow cytometric analysis for the comparison of the original _Snrpn_-EGFP clone (original clone) with the GT-A (top) and GT-B (bottom) clones. (C) Schematic representations of the trapped genes. (D) Expression analysis of Arid4a, Arid4b, and their transcripts fused with _β_geo by RT–PCR. “e” refers to exon number, as in e1 for exon 1. (E) Methylation analysis of the Snrpn CpG island in the original _Snrpn_-EGFP clone and three gene trap clones. Southern blot analysis is similar to Figure 1D.
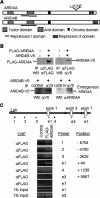
FIGURE 3.
Interaction of ARID4A with ARID4B and with the Snrpn promoter. (A) Comparison of ARID4A and ARID4B. (B) Association between ARID4A and ARID4B. (Top) Expression vectors for Flag-ARID4A and ARID4B-V5 were cotransfected into 293 cells. Immunoprecipitation was performed with anti-Flag antibody. Total ARID4A and coimmunoprecipitated ARID4B were detected by Western blot assays using anti-Flag antibody (left) and anti-V5 antibody (right), respectively. (Bottom) 293 cells were transfected with the ARID4B-V5 vector. Immunoprecipitation was performed with anti-V5 antibody and normal mouse IgG as a control antibody. Total ARID4B and coimmunoprecipitated cell-endogenous ARID4A were detected by anti-V5 antibody (left) and an antibody for ARID4A (right), respectively. (C) ChIP analysis for ARID4A in the Snrpn promoter region. (Top) The Snrpn promoter region is shown with bars to indicate the amplified region of each PCR product used in the ChIP analysis. The amplified products from primer sets of 1, 2, 3, e1, and 4 were within or near the Snrpn promoter. Primer sets e1, e3, and e7 overlap the exons of the respective numbers. (Bottom) Cross-linked chromatin was immunoprecipitated with anti-Flag antibody from wild-type ES cells (control) or the ES clone stably expressing Flag-tagged ARID4A. DNA from immunoprecipitated chromatin or cell extracts (1% input) was subjected to PCR for the sites shown above.
FIGURE 4.
Targeted disruption of the Arid4a and Arid4b genes. (A) Targeting strategy for disrupting the Arid4a gene in ES cells. (IRES) Internal ribosome entry site; (TK) herpes simplex viral thymidine kinase cassette; (E) EcoRI; (N) NcoI; (X) XhoI. (B) Southern blot analysis of the Arid4a targeted ES clone using the 5′-flanking probe (top) and the 3′-flanking probe (bottom). (Wt) Wild-type allele; (mt) mutant allele. (C) Southern blotting using the 3′-flanking probe for genotype analysis of offspring from the Arid4a +/− mice intercross. (D) RT–PCR analysis for Arid4a expression using primer pairs in exons 1 and 5 (e1-e5) in wild-type, Arid4a +/−, or Arid4a −/− mice. (E) Targeting strategy for disrupting the Arid4b gene in ES cells. Horizontal bold arrows (a, b, and c) indicate the positions of primers used to assay genotype in F; primers a and b amplify the wild-type allele, and b and c amplify the mutant allele. (S) SstI; (E) EcoRI; (H) HindIII; (P) PstI. (F) Genotype analysis of the _Arid4b_-targeted ES clone by Southern blotting using the 5′-flanking probe (top) and by PCR using the primers shown in E (bottom). (G) Genotype identification of a litter of six E3.5 embryos from mating between Arid4b +/− heterozygotes by PCR as in F.
FIGURE 5.
Epigenetic analysis of the PWS-IC in Arid4a −/− Arid4b +/− mice. (A) Southern blot analysis for DNA methylation at the Snrpn CpG island. Genomic DNA isolated from seven Arid4a −/− Arid4b +/− mice and one wild-type control was digested with HindIII (H) alone or in combination with SacII (HS) for Southern blot analysis. (Me) The 14-kb methylated DNA; (unme) the 10.2-kb unmethylated DNA from the Snrpn allele. (B) Bisulfite sequence analysis of methylation status of 16 CpG dinucleotides across the exon 1 region (−307 to +115) of Snrpn in wild-type mice and Arid4a −/− Arid4b +/− mice (mutant mice 1–5). Each line represents an individual clone with open and closed circles corresponding to unmethylated and methylated CpGs, respectively. (S) A SacII site; (DMR) differential methylation region. (C) ChIP analysis for H3K9 and H4K20 trimethylation at the Snrpn promoter in the Arid4a −/− Arid4b +/− mice (mutant mice 1 and 5). (Top) DNA isolated from immunoprecipitated chromatin using anti-trimethylated H3K9 or anti-trimethylated H4K20 antibodies and from cell crude extracts (2% input) was subjected to PCR using the primer pair e1 shown in Figure 3C. (Bottom) Quantification of trimethylated H3K9 and trimethylated H4K20 by real-time PCR analysis. (D) ChIP analysis for wild-type mice, mice paternally inheriting the 4.8-kb deletion (Δ4.8 m+/p−), or mice maternally inheriting the 4.8-kb deletion (Δ4.8 m−/p+) using anti-trimethylated H3K9 (top) or anti-trimethylated H4K20 (bottom) antibodies. Quantification of trimethylated H3K9 and trimethylated H4K20 in the Snrpn promoter region was assayed by real-time PCR using the primer pair e1 shown in Figure 3C. (E) Quantitative RT–PCR analysis for two paternally expressed genes, Snrpn (left) and Ndn (right), in wild-type mice and the Arid4a −/− Arid4b +/− mice. The level of gene expression measured for the wild-type mice was set as 1. (F) Western blot analysis for maternally expressed gene Ube3a using an antibody to E6-AP or a control antibody to actin. Ratios of expression were quantitated by densitometry. The level of E6-AP was normalized against actin level for each sample. The level of Ube3a expression measured for the wild-type mouse was set as 1.
FIGURE 6.
Suppression of an AS-IC imprinting defect by the Arid4a and Arid4b mutations. (A) Southern blot analysis for methylation patterns at the Snrpn and Ndn CpG islands in progeny from mating a female bearing the AS-ICan mutation to a male carrying the Arid4b mutation. Genomic DNA was digested with HindIII (H) alone or in combination with SacII (HS) and hybridized with a probe from Snrpn intron 1 (top) or from the Ndn 5′-flanking region (bottom). Fragment sizes for the Snrpn CpG island: me, 14 kb; unme, 10.2 kb. Fragment sizes for the Ndn CpG island: me, 3.4 kb; unme, 1.9 kb. (me) Methylated; (unme) unmethylated. (B–D) Summary of methylation status at Snrpn in progeny with all genotypes from mating female mice bearing the AS-ICan mutation to male mice carrying the Arid4b +/− (B), Arid4a −/− (C), or Rb +/− (D) mutations. (E) Expression analysis for Ube3a. Protein was extracted from the cerebral cortex of mice with different mutation combinations accompanied by either differential methylation (DM) or fully unmethylated (UM) at Snrpn. Western blot analysis was performed using an antibody to E6-AP or to actin.
FIGURE 7.
Completed suppression of the AS-IC imprinting defect. In the first generation, female mice with the AS-ICan mutation were mated to male mice homozygous for the Arid4a mutation. In the second generation, double-heterozygous females with a lack of methylation at Snrpn were mated to Arid4a −/− males (A) and wild-type males (B), or double-heterozygous females with differential methylation at Snrpn were mated to wild-type males (C). The methylation status of Snrpn was analyzed in progeny with all four genotypes in the third generation.
Similar articles
- Identification of chromatin remodeling genes Arid4a and Arid4b as leukemia suppressor genes.
Wu MY, Eldin KW, Beaudet AL. Wu MY, et al. J Natl Cancer Inst. 2008 Sep 3;100(17):1247-59. doi: 10.1093/jnci/djn253. Epub 2008 Aug 26. J Natl Cancer Inst. 2008. PMID: 18728284 Free PMC article. - Influence of the Prader-Willi syndrome imprinting center on the DNA methylation landscape in the mouse brain.
Brant JO, Riva A, Resnick JL, Yang TP. Brant JO, et al. Epigenetics. 2014 Nov;9(11):1540-56. doi: 10.4161/15592294.2014.969667. Epigenetics. 2014. PMID: 25482058 Free PMC article. - An unexpected function of the Prader-Willi syndrome imprinting center in maternal imprinting in mice.
Wu MY, Jiang M, Zhai X, Beaudet AL, Wu RC. Wu MY, et al. PLoS One. 2012;7(4):e34348. doi: 10.1371/journal.pone.0034348. Epub 2012 Apr 4. PLoS One. 2012. PMID: 22496793 Free PMC article. - Imprinting defects on human chromosome 15.
Horsthemke B, Buiting K. Horsthemke B, et al. Cytogenet Genome Res. 2006;113(1-4):292-9. doi: 10.1159/000090844. Cytogenet Genome Res. 2006. PMID: 16575192 Review. - Structure and function of the human chromosome 15 imprinting center.
Horsthemke B. Horsthemke B. J Cell Physiol. 1997 Nov;173(2):237-41. doi: 10.1002/(SICI)1097-4652(199711)173:2<237::AID-JCP28>3.0.CO;2-B. J Cell Physiol. 1997. PMID: 9365529 Review.
Cited by
- Heterozygous Knockout of ARID4B Using CRISPR/Cas9 Attenuates Some Aggressive Phenotypes in a Breast Cancer Cell Line.
Gonzalez-Salinas F, Herrera-Gamboa J, Rojo R, Trevino V. Gonzalez-Salinas F, et al. Genes (Basel). 2023 Dec 6;14(12):2184. doi: 10.3390/genes14122184. Genes (Basel). 2023. PMID: 38137006 Free PMC article. - Genome-wide patterns of genetic variation in two domestic chickens.
Fan WL, Ng CS, Chen CF, Lu MY, Chen YH, Liu CJ, Wu SM, Chen CK, Chen JJ, Mao CT, Lai YT, Lo WS, Chang WH, Li WH. Fan WL, et al. Genome Biol Evol. 2013;5(7):1376-92. doi: 10.1093/gbe/evt097. Genome Biol Evol. 2013. PMID: 23814129 Free PMC article. - Temporal-Spatial Establishment of Initial Niche for the Primary Spermatogonial Stem Cell Formation Is Determined by an ARID4B Regulatory Network.
Wu RC, Zeng Y, Chen YF, Lanz RB, Wu MY. Wu RC, et al. Stem Cells. 2017 Jun;35(6):1554-1565. doi: 10.1002/stem.2597. Epub 2017 Mar 16. Stem Cells. 2017. PMID: 28207192 Free PMC article. - Recommendations for the investigation of animal models of Prader-Willi syndrome.
Resnick JL, Nicholls RD, Wevrick R; Prader-Willi Syndrome Animal Models Working Group. Resnick JL, et al. Mamm Genome. 2013 Jun;24(5-6):165-78. doi: 10.1007/s00335-013-9454-2. Epub 2013 Apr 23. Mamm Genome. 2013. PMID: 23609791 - Identification of the PTEN-ARID4B-PI3K pathway reveals the dependency on ARID4B by PTEN-deficient prostate cancer.
Wu RC, Young IC, Chen YF, Chuang ST, Toubaji A, Wu MY. Wu RC, et al. Nat Commun. 2019 Sep 24;10(1):4332. doi: 10.1038/s41467-019-12184-8. Nat Commun. 2019. PMID: 31551414 Free PMC article.
References
- Bannister, A.J., Kouzarides, T. Reversing histone methylation. Nature. 2005;436:1103–1106. - PubMed
- Bannister, A.J., Zegerman, P., Partridge, J.F., Miska, E.A., Thomas, J.O., Allshire, R.C., Kouzarides, T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. - PubMed
- Bressler, J., Tsai, T.F., Wu, M.Y., Tsai, S.F., Ramirez, M.A., Armstrong, D., Beaudet, A.L. The SNRPN promoter is not required for genomic imprinting of the Prader-Willi/Angelman domain in mice. Nat. Genet. 2001;28:232–240. - PubMed
- Buiting, K., Saitoh, S., Gross, S., Dittrich, B., Schwartz, S., Nicholls, R.D., Horsthemke, B. Inherited micro-deletions in the Angelman and Prader-Willi syndromes define an imprinting centre on human chromosome 15. Nat. Genet. 1995;9:395–400. - PubMed
- Buiting, K., Lich, C., Cottrell, S., Barnicoat, A., Horsthemke, B. A 5-kb imprinting center deletion in a family with Angelman syndrome reduces the shortest region of deletion overlap to 880 bp. Hum. Genet. 1999;105:665–666. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous