Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer - PubMed (original) (raw)
Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer
Julie Huang et al. Genes Dev. 2006.
Abstract
Silencing within the yeast ribosomal DNA (rDNA) repeats protects the integrity of this highly repetitive array by inhibiting hyperrecombination and repressing transcription from foreign promoters. Using affinity purification combined with highly sensitive mixture mass spectrometry, we have analyzed the protein interaction network involved in suppressing homologous recombination within the rDNA locus. We show that the Net1 and Sir2 subunits of the RENT (regulator of nucleolar silencing and telophase exit) silencing complex, and Fob1, which recruits RENT to the nontranscribed spacer I (NTS1) region of rDNA, are physically associated with Tof2. In addition to RENT components and Fob1, Tof2 copurified with a two-subunit complex composed of Lrs4 and Csm1. Tof2, Lrs4, and Csm1 are recruited to the NTS1 region by Fob1 and are specifically required for silencing at this rDNA region. Moreover, Lrs4 and Csm1 act synergistically with Sir2 to suppress unequal crossover at the rDNA and are released from the nucleolus during anaphase. Together with previous observations showing that Csm1 physically associates with cohesin, these findings suggest a possible model in which RENT, Tof2, and Lrs4/Csm1 physically clamp rDNA to the cohesin ring, thereby restricting the movement of rDNA sister chromatids relative to each other to inhibit unequal exchange.
Figures
Figure 1.
Affinity purifications of rDNA silencing complexes. Silver-stained SDS-PAGE gels of complexes purified from untagged (A), Net1-TAP (B), Sir2-TAP (C), and Fob1-TAP (D) cells; 2.5% of the total purified material is shown. (E) The results of the total mixture analysis by liquid chromatography combined with tandem mass spectrometry (LC-MS/MS). (F) The protein sequence alignment of the N termini of Tof2 and Net1 indicates 30% identity and 53% similarity within the N-terminal 250 amino acids (shaded area). (CBP) Calmodulin-binding protein.
Figure 2.
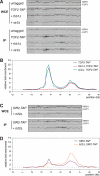
Tof 2 is a nucleolar protein required specifically for rDNA silencing at NTS1. (A) The physical structure of the tandemly repeating RDN1 locus of S. cerevisiae is shown above, and a single 9.1-kb rDNA unit is shown expanded below. Each repeat yields a Pol I-transcribed 35S precursor rRNA (shown as a divided thick arrow) and a Pol III-transcribed 5S rRNA (arrowhead). The 35S coding regions are separated by an NTS, which is divided by the 5S gene into NTS1 and NTS2. Solid bars indicate the recombination enhancer (RE) region and the Pol I TIR. The locations of the RFB () and autonomously replicating sequences () are indicated. Vertical arrows indicate insertion sites of the NTS1 and NTS2/TIR silencing reporters. (B) TOF 2 is required for rDNA silencing at NTS1 but not at NTS2/TIR. Silencing within rDNA was assessed by monitoring the growth of 10-fold serial dilutions of cells plated on−URA medium. SC medium was used as a plating control. TOF 2 is required specifically for silencing only at NTS1, unlike SIR2, which is required for silencing at both NTS1 and NTS2. Locations of rDNA reporter genes are as indicated in Figure 2A. (C) TOF 2 and FOB1 are not required for telomeric silencing. Silencing was assessed by monitoring the growth of 10-fold serial dilutions of cells on SC (synthetic complete) medium supplemented with 5-FOA. SC medium was used as a plating control. The URA3 reporter gene was integrated either adjacent to the telomeric repeats of Chromosome VIIL (TELVIIL)or ~15 kb away, at the ADH4 locus. (D) Tof2 colocalizes with nucleolar marker Net1. Immunofluorescence images show the subcellular localization of Net1-GFP (green), Tof2-HA3 (red), and DAPI-stained DNA (blue). The merged image shows that Net1-GFP and Tof2-HA3 colocalize to nucleolar domains that are nonoverlapping with the rest of the genome (yellow). Arrows indicate dividing cells.
Figure 3.
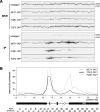
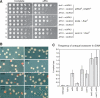
Tof2 associates primarily with the NTS1 region of rDNA. (A) PCR products amplified from WCE (upper panels) and IP (lower panels) chromatin. Multiplex PCR was performed to amplify RDN1 and CUP1 sequences as indicated. PCR products 1–4 and 6–34 are shown. (B) Representative graph showing the relative fold association of Tof2-TAP (solid black line), Net1-TAP (dashed gray line), and Fob1-TAP (solid gray line) across the rDNA repeat. A schematic representation of the rDNA unit is shown below the graph, with significant features shown as in Figure 2A and PCR products depicted below. Most of the Tof2-TAP is concentrated within NTS1, with a smaller peak observed near the border of NTS2 and the 35S rRNA coding region.
Figure 4.
Tof2-TAP requires FOB1 but not SIR2 for association with NTS1. (A,C) Examples of ChIP data showing PCR products amplified from WCE and IP DNA. Multiplex PCR was performed to amplify RDN1 and CUP1 sequences as indicated. PCR products 4 and 6–33 are shown. (B) Representative graphs showing the association of Tof2-TAP across an rDNA repeat in wild-type (blue), _fob1_Δ (red), or _sir2_Δ (green) cells. (D) Sir2-TAP association with rDNA is largely Tof2 independent.
Figure 5.
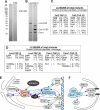
Affinity purifications of native Tof2, Lrs4, and Csm1 complexes. Silver-stained SDS-PAGE gels of native complexes purified from Tof2-TAP (A) and Lrs4-TAP and Csm1-TAP (B); 2.5% of total purified material is shown. (C,D) The results of the total mixture analysis by liquid chromatography combined with tandem mass spectrometry (LC-MS/MS). (E,F) Summaries of the protein–protein interaction network of silencing factors. Arrows indicate physical interactions determined by affinity purifications from this work (blue) or by others (green), or by yeast two-hybrid (red). Direction of the arrowheads is from bait toward interactor.
Figure 6.
Lrs4-TAP and Csm1-TAP associate with NTS1 in an FOB1- and _TOF 2_-dependent manner. Examples of ChIP data showing PCR products amplified from WCE and IP DNA associated with Lrs4-TAP (A) and Csm1-TAP (C). Multiplex PCR was performed to amplify RDN1 and CUP1 sequences as indicated. PCR products 2–4, 6–34, and 1 (A) and 10–18 and 23 (C) are shown. Quantifications of these data are shown in B and D. Both Lrs4-TAP and Csm1-TAP associate significantly with NTS1 but not with the NTS2/35S region.
Figure 7.
LRS4 and CSM1 are required for NTS1 silencing and unequal sister chromatid exchange. (A) Silencing was assayed as described in Figure 2B, and locations of reporters are shown in Figure 2A. Cells lacking LRS4 or CSM1 exhibit a complete loss of silencing at NTS1 but wild-type levels of silencing at NTS2. (B) Unequal sister chromatid exchange is monitored by loss of the ADE2 gene located within the rDNA array. Cells expressing ADE2 are white, while cells lacking the ADE2 gene are red. Half-sectored colonies represent loss of the marker during the first division upon plating. Entirely red colonies are descended from a cell that has lost the marker prior to plating. (C) Unequal sister chromatid exchange is represented as percent marker loss, calculated as the ratio of half-sectored colonies to the total number of colonies, excluding entirely red colonies.
Figure 8.
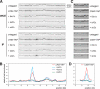
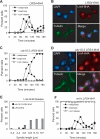
Lrs4-6HA is released from the nucleolus during anaphase. (A) Wild-type cells (A13838) carrying an LRS4-6HA fusion were arrested in G1 in YEPD medium with α-factor (5 μg/mL). When arrest was complete, cells were released into YEPD medium lacking pheromone at 23°C. At the indicated times, samples were taken to determine the percentage of cells with metaphase and anaphase spindles, as well as the percentage of cells with Lrs4-6HA released from the nucleolus. (B) An example of Lrs4 release in anaphase cells. Lrs4-6HA is shown in red, microtubules in green, and DNA in blue. (C) cdc15-2 cells (A13839) carrying an LRS4-6HA fusion were arrested in G1 in YEPD medium with α-factor (5 μg/mL). When arrest was complete, cells were released into YEPD medium lacking pheromone at 37°C. At the indicated times, samples were taken to determine the percentages of cells with metaphase and anaphase spindles and the percentage of cells with Lrs4-6HA released from the nucleolus. (D) An example of Lrs4 localization in cdc15-2 cells. Lrs4 is shown in red, microtubules in green, and DNA in blue. (E) Wild-type (A13838) and cdc15-2 cells (A13839) were grown as described in A, and the localization of Lrs4-6HA was determined with respect to the length of the mitotic spindle as described in Stegmeier et al. (2004). (F) Wild-type (A13838) and _net1_Δ cells (A14568) were grown as described in A. At the indicated times, samples were taken to determine the percentages of cells with metaphase and anaphase spindles, and the percentage of cells with Lrs4-6HA released from the nucleolus.
Figure 9.
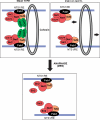
Model for a protein bridge that inhibits recombination by unequal crossover. Within NTS1, RE sequences are bound by Fob1, which is required for the recruitment of the RENT complex, consisting of Net1, Sir2, and Cdc14. Fob1 also recruits Tof2, which is required for the association of Lrs4/ Csm1 with RE sequences. Lrs4/Csm1 may form a protein bridge that clamps sister chromatids together, either directly or through association with cohesin. Lrs4/Csm1–cohesin association would clamp rDNA to the cohesin ring, thereby restricting the movement of sister chromatids relative to each other to inhibit unequal exchange. (Bottom) The bridge is disassembled during mitosis by the release of cohesin and Lrs4/Csm1. (Right side) In _lrs4_Δ and _csm1_Δ mutant cells, cohesin is no longer clamped to rDNA, allowing unrestricted movement of sister chromatids relative to each other.
Similar articles
- Yeast histone H3 lysine 4 demethylase Jhd2 regulates mitotic rDNA condensation.
Ryu HY, Ahn S. Ryu HY, et al. BMC Biol. 2014 Sep 24;12:75. doi: 10.1186/s12915-014-0075-3. BMC Biol. 2014. PMID: 25248920 Free PMC article. - Mechanism of Regulation of Intrachromatid Recombination and Long-Range Chromosome Interactions in Saccharomyces cerevisiae.
Zaman S, Choudhury M, Jiang JC, Srivastava P, Mohanty BK, Danielson C, Humphrey SJ, Jazwinski SM, Bastia D. Zaman S, et al. Mol Cell Biol. 2016 May 2;36(10):1451-63. doi: 10.1128/MCB.01100-15. Print 2016 May 15. Mol Cell Biol. 2016. PMID: 26951198 Free PMC article. - Regulation of rDNA stability by sumoylation.
Eckert-Boulet N, Lisby M. Eckert-Boulet N, et al. DNA Repair (Amst). 2009 Apr 5;8(4):507-16. doi: 10.1016/j.dnarep.2009.01.015. Epub 2009 Mar 3. DNA Repair (Amst). 2009. PMID: 19261548 Review. - Nucleolus in the spotlight.
Hernandez-Verdun D. Hernandez-Verdun D. Cell Cycle. 2005 Jan;4(1):106-8. doi: 10.4161/cc.4.1.1355. Epub 2005 Jan 10. Cell Cycle. 2005. PMID: 15611637 Review.
Cited by
- The Epigenetic Pathways to Ribosomal DNA Silencing.
Srivastava R, Srivastava R, Ahn SH. Srivastava R, et al. Microbiol Mol Biol Rev. 2016 Jun 1;80(3):545-63. doi: 10.1128/MMBR.00005-16. Print 2016 Sep. Microbiol Mol Biol Rev. 2016. PMID: 27250769 Free PMC article. Review. - The Reb1-homologue Ydr026c/Nsi1 is required for efficient RNA polymerase I termination in yeast.
Reiter A, Hamperl S, Seitz H, Merkl P, Perez-Fernandez J, Williams L, Gerber J, Németh A, Léger I, Gadal O, Milkereit P, Griesenbeck J, Tschochner H. Reiter A, et al. EMBO J. 2012 Aug 15;31(16):3480-93. doi: 10.1038/emboj.2012.185. Epub 2012 Jul 17. EMBO J. 2012. PMID: 22805593 Free PMC article. - Interference between DNA replication and transcription as a cause of genomic instability.
Lin YL, Pasero P. Lin YL, et al. Curr Genomics. 2012 Mar;13(1):65-73. doi: 10.2174/138920212799034767. Curr Genomics. 2012. PMID: 22942676 Free PMC article. - The Lrs4-Csm1 monopolin complex associates with kinetochores during anaphase and is required for accurate chromosome segregation.
Brito IL, Monje-Casas F, Amon A. Brito IL, et al. Cell Cycle. 2010 Sep 1;9(17):3611-8. doi: 10.4161/cc.9.17.12885. Epub 2010 Sep 1. Cell Cycle. 2010. PMID: 20818155 Free PMC article. - Yeast sirtuins and the regulation of aging.
Wierman MB, Smith JS. Wierman MB, et al. FEMS Yeast Res. 2014 Feb;14(1):73-88. doi: 10.1111/1567-1364.12115. Epub 2013 Nov 14. FEMS Yeast Res. 2014. PMID: 24164855 Free PMC article. Review.
References
- Aparicio, O.M., Billington, B.L., Gottschling, D.E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae . Cell. 1991;66:1279–1287. - PubMed
- Bernard, P., Maure, J.F., Partridge, J.F., Genier, S., Javerzat, J.P., Allshire, R.C. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. - PubMed
- Brewer, B.J., Fangman, W.L. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. - PubMed
- Bryk, M., Banerjee, M., Murphy, M., Knudsen, K.E., Garfinkel, D.J., Curcio, M.J. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes & Dev. 1997;11:255–269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM061641/GM/NIGMS NIH HHS/United States
- R01 GM061641/GM/NIGMS NIH HHS/United States
- R29 GM056800/GM/NIGMS NIH HHS/United States
- R01 GM056800/GM/NIGMS NIH HHS/United States
- GM56800/GM/NIGMS NIH HHS/United States
- R01 GM067945/GM/NIGMS NIH HHS/United States
- GM67945/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases