Ancient polymorphism and functional variation in the primate MHC-DQA1 5' cis-regulatory region - PubMed (original) (raw)
Ancient polymorphism and functional variation in the primate MHC-DQA1 5' cis-regulatory region
Dagan A Loisel et al. Proc Natl Acad Sci U S A. 2006.
Abstract
Precise regulation of MHC gene expression is critical to vertebrate immune surveillance and response. Polymorphisms in the 5' proximal promoter region of the human class II gene HLA-DQA1 have been shown to influence its transcriptional regulation and may contribute to the pathogenesis of autoimmune diseases. We investigated the evolutionary history of this cis-regulatory region by sequencing the DQA1 5' proximal promoter region in eight nonhuman primate species. We observed unexpectedly high levels of sequence variation and multiple strong signatures of balancing selection in this region. Specifically, the considerable DQA1 promoter region diversity was characterized by abundant shared (or trans-species) polymorphism and a pronounced lack of fixed differences between species. The majority of transcription factor binding sites in the DQA1 promoter region were polymorphic within species, and these binding site polymorphisms were commonly shared among multiple species despite evidence for negative selection eliminating a significant fraction of binding site mutations. We assessed the functional consequences of intraspecific promoter region diversity using a cell line-based reporter assay and detected significant differences among baboon DQA1 promoter haplotypes in their ability to drive transcription in vitro. The functional differentiation of baboon promoter haplotypes, together with the significant deviations from neutral sequence evolution, suggests a role for balancing selection in the evolution of DQA1 transcriptional regulation in primates.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
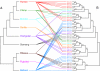
Fig. 1.
Incongruity of primate species tree and DQA1 promoter region gene tree. (A) Species tree of the nine species from which DQA1 promoter sequences were obtained. (B) The most probable DQA1 promoter tree topology from the Bayesian analysis, with posterior probabilities of branch support. Branch lengths for both trees are arbitrary. Species and gene tree were reconciled by using Page's method (38) for reconciling phylogenetic trees (69).
Fig. 2.
Transcription factor binding site motifs in the DQA1 proximal promoter in primates. Letters A–H refer to unique transcription factor binding site motifs, with the corresponding sequence represented above. W, NF-κB, S, J, X1, X2, and Y refer to binding sites identified in the DQA1 promoter (3, 11, 12). Branch lengths of the species phylogeny on the left (27, 28) are roughly proportional to the timeline indicated below it. MYA, million years ago.
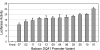
Fig. 3.
Luciferase activity induced by baboon DQA1 promoter haplotypes in a transient transfection reporter assay. Bars represent luciferase activity of 12 different baboon DQA1 promoter constructs calculated as the ratio of firefly to Renilla (coreporter) luminescence. Error bars represent standard error of the mean estimated from three replicate experiments. Numbers on the x axis correspond to the unique baboon DQA1 promoter haplotypes, and “Empty” refers to the promoter-less pGL3 basic reporter construct.
Similar articles
- Conserved residues of the bare lymphocyte syndrome transcription factor RFXAP determine coordinate MHC class II expression.
Long AB, Ferguson AM, Majumder P, Nagarajan UM, Boss JM. Long AB, et al. Mol Immunol. 2006 Feb;43(5):395-409. doi: 10.1016/j.molimm.2005.03.008. Epub 2005 Apr 12. Mol Immunol. 2006. PMID: 16337482 - HLA-DQA1 and HLA-DQB1 alleles and haplotypes in two Brazilian Indian tribes: evidence of conservative evolution of HLA-DQ.
Sotomaior VS, Faucz FR, Schafhauser C, Janzen-Dück M, Boldt AB, Petzl-Erler ML. Sotomaior VS, et al. Hum Biol. 1998 Aug;70(4):789-97. Hum Biol. 1998. PMID: 9686487 - [Association of polymorphism for HLA-DQA1 promotor region (QAP) with IDDM].
Qiu C, Song C, Zhou W, Hu X, Bai S. Qiu C, et al. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 1994 Dec;16(6):411-5. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 1994. PMID: 7720136 Chinese. - Differential transcription elements direct expression of HLA-DQ genes.
Woolfrey AE, Nepom GT. Woolfrey AE, et al. Clin Immunol Immunopathol. 1995 Feb;74(2):119-26. doi: 10.1006/clin.1995.1018. Clin Immunol Immunopathol. 1995. PMID: 7828365 Review. - Regulatory mutations and human genetic disease.
Cooper DN. Cooper DN. Ann Med. 1992 Dec;24(6):427-37. doi: 10.3109/07853899209166991. Ann Med. 1992. PMID: 1283065 Review.
Cited by
- HLA-DRB1 genes and the expression dynamics of HLA CIITA determine the susceptibility to T2DM.
Chinniah R, Sevak V, Pandi S, Ravi PM, Vijayan M, Kannan A, Karuppiah B. Chinniah R, et al. Immunogenetics. 2021 Aug;73(4):291-305. doi: 10.1007/s00251-021-01212-x. Epub 2021 Mar 22. Immunogenetics. 2021. PMID: 33754173 - Human MHC architecture and evolution: implications for disease association studies.
Traherne JA. Traherne JA. Int J Immunogenet. 2008 Jun;35(3):179-92. doi: 10.1111/j.1744-313X.2008.00765.x. Epub 2008 Apr 8. Int J Immunogenet. 2008. PMID: 18397301 Free PMC article. Review. - Evidence that purifying selection acts on promoter sequences.
Arthur RK, Ruvinsky I. Arthur RK, et al. Genetics. 2011 Nov;189(3):1121-6. doi: 10.1534/genetics.111.133637. Epub 2011 Sep 6. Genetics. 2011. PMID: 21900262 Free PMC article. - Balancing selection is common in the extended MHC region but most alleles with opposite risk profile for autoimmune diseases are neutrally evolving.
Cagliani R, Riva S, Pozzoli U, Fumagalli M, Comi GP, Bresolin N, Clerici M, Sironi M. Cagliani R, et al. BMC Evol Biol. 2011 Jun 17;11:171. doi: 10.1186/1471-2148-11-171. BMC Evol Biol. 2011. PMID: 21682861 Free PMC article. - Genomic analysis of Ovis aries (Ovar) MHC class IIa loci.
Herrmann-Hoesing LM, White SN, Kappmeyer LS, Herndon DR, Knowles DP. Herrmann-Hoesing LM, et al. Immunogenetics. 2008 Apr;60(3-4):167-76. doi: 10.1007/s00251-008-0275-5. Epub 2008 Mar 6. Immunogenetics. 2008. PMID: 18322680
References
- Davidson EH. Genomic Regulatory Systems: Development and Evolution. San Diego: Academic; 2001.
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. Mol Biol Evol. 2003;20:1377–1419. - PubMed
- Rohn WM, Lee YJ, Benveniste EN. Crit Rev Immunol. 1996;16:311–330. - PubMed
- Ting JP, Trowsdale J. Cell. 2002;109:S21–S33. - PubMed
- Apanius V, Penn D, Slev PR, Ruff LR, Potts WK. Crit Rev Immunol. 1997;17:179–224. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials