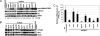A pathogen-inducible endogenous siRNA in plant immunity - PubMed (original) (raw)
A pathogen-inducible endogenous siRNA in plant immunity
Surekha Katiyar-Agarwal et al. Proc Natl Acad Sci U S A. 2006.
Abstract
RNA interference, mediated by small interfering RNAs (siRNAs), is a conserved regulatory process that has evolved as an antiviral defense mechanism in plants and animals. It is not known whether host cells also use siRNAs as an antibacterial defense mechanism in eukaryotes. Here, we report the discovery of an endogenous siRNA, nat-siRNAATGB2, that is specifically induced by the bacterial pathogen Pseudomonas syringae carrying effector avrRpt2. We demonstrate that the biogenesis of this siRNA requires DCL1, HYL1, HEN1, RDR6, NRPD1A, and SGS3. Its induction also depends on the cognate host disease resistance gene RPS2 and the NDR1 gene that is required for RPS2-specified resistance. This siRNA contributes to RPS2-mediated race-specific disease resistance by repressing PPRL, a putative negative regulator of the RPS2 resistance pathway.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
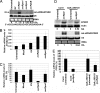
Fig. 1.
A nat-siRNA is induced by Pst (avrRpt2). (A) Detection of the nat-siRNA by Northern blot analysis. The nat-siRNA sequence is shown under the panel. Small RNA was extracted from the leaves harvested at 15 hpi of Pst (2 × 107 cfu/ml) carrying various avr genes and an oligonucleotide probe complementary to the siRNA was used. DNA probe was used for detecting U6 RNA for measuring the relative abundance (RA) (shown below). Ethidium bromide-stained tRNA is also shown as a loading control. (B and C) Relative expression levels of ATGB2 (1B) and PPRL (1C) as measured by real-time RT-PCR. The expression levels of AtGB2 and PPRL were normalized to that of ubiquitin. (D) Northern blot analysis shows reduced ATGB2 and nat-siRNAATGB2 expression levels in Salk_083103 homozygous line upon Pst (avrRpt2) (2 × 107 cfu/ml) challenge comparing to the WT Col-0. The levels of actin and U6 were used for quantification and loading controls. (E) Relative mRNA level of PPRL in Salk_083103 homozygous line. The expression levels in untreated WT Col-0 were used as 100 % and standard deviations were plotted from three replicates (B, C, and E).
Fig. 2.
Accumulation of nat-siRNAATGB2 depends on DCL1, HYL1, and RDR6, and also requires HEN1, NRPD1a, and SGS3. (A) Northern blot analysis of nat-siRNAATGB2 in various Pst (avrRpt2)-treated small RNA biogenesis mutants and their corresponding WT controls. MiR171 and U6 RNA was used as controls. U6 level was used for quantification. (B) Relative PPRL mRNA levels in sgs3, _dcl1_-9, hyl1, rdr6, nrpd1a, and their corresponding WT controls after Pst (avrRpt2) infection. The expression levels were normalized to that of ubiquitin. The expression level in untreated WT Col-0 was used as 100%. Standard deviations were plotted from three replicates. (C) Transient coexpression of PPRL and ATGB2 in N. benthamiana. Agrobacterium GV3101 harboring PPRL was coinfiltrated with GV3101 carrying full-length (F) or only the overlapping region (O) of ATGB2 constructs into 3-week-old N. benthamiana leaves. The expression of AtGB2 was induced by Dex at 2 dpi, and tissue was harvested after 24 h of induction. The expression of PPRL was measured at both RNA and protein levels by semiquantitative RT-PCR (Top) and Western blot analysis (Sigma anti-FLAG, 1:2,000 dilution, Middle), respectively. Actin was used as a control for RT-PCR. The Rubisco large subunit from a gel that was run in parallel was stained with Coomassie blue for Western blot loading control. nat-siRNAATGB2 is detected after the induction of either full-length or overlapping region of ATGB2. (Bottom) tRNA was used as a loading control.
Fig. 3.
Accumulation of nat-siRNAATGB2 is controlled by RPS2 and some components of the disease resistance signaling pathway. Northern blot analysis of nat-siRNAATGB2 (A) and ATGB2 (B) was performed on Pst (avrRpt2)-treated defense-signaling mutants and WT Col-0 plants. U6 RNA was used for small RNA quantification. (C) Relative quantification of PPRL expression in defense signaling mutants by real-time RT-PCR analysis. PPRL expression level was normalized to that of ubiquitin. The expression levels in untreated WT Col-0 were used as 100%. Standard deviations were plotted from three replicates.
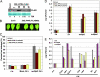
Fig. 4.
Overexpression of PPRL attenuates _RPS2_-mediated resistance in Arabidopsis plants. (A) Western blot analysis of transgenic Arabidopsis plants overexpressing PPRL (Sigma anti-FLAG, 1:2,000 dilution). Shown is the Rubisco large subunit from a gel that was run in parallel and stained with Coomassie blue. Two lines with high (line 32) or low (line 33) expression level of PPRL were selected for phenotypic analysis. (B) PPRL overexpression line displays delayed HR. Picture of line 32 was taken at 16 hpi of Pst (avrRpt2) (2 × 107 cfu/ml). (C) PPRL overexpression lines exhibit reduced electrolyte leakage. Plants treated with 10 mM MgCl2 and Pst (avrRpt2) (1 × 107 cfu/ml) were measured at 0 and 24 hpi. Error bars represent standard deviation of four replicates. Similar results were obtained from two independent experiments. (D) PPRL overexpression lines display enhanced pathogen growth of Pst (avrRpt2). Bacterial growth was measured at 0 and 4 dpi of Pst carrying EV, avrRpt2 or avrRpm1 (2 × 105 cfu/ml). Error bars represent standard deviation of five replicates. Similar results were obtained in three independent experiments. (E) Pst (avrRpt2) accumulates to a higher level in rdr6 and hyl1, but not in dcl3, as compared with that in the corresponding WT C24, No, or Col-0, respectively. No difference was observed in the growth of Pst (avrRpm1) between the mutants rdr6, hyl1, or dcl3 and their corresponding WT control. Pathogen growth was measured at 0 and 4 dpi. Similar results were obtained in two independent experiments.
Fig. 5.
Model for nat-siRNAATGB2 biogenesis and function. Components in red are required for nat-siRNAATGB2 formation. RISC, RNA-induced silencing complex.
Similar articles
- A key role for the Arabidopsis WIN3 protein in disease resistance triggered by Pseudomonas syringae that secrete AvrRpt2.
Lee MW, Lu H, Jung HW, Greenberg JT. Lee MW, et al. Mol Plant Microbe Interact. 2007 Oct;20(10):1192-200. doi: 10.1094/MPMI-20-10-1192. Mol Plant Microbe Interact. 2007. PMID: 17918621 - Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins.
Moissiard G, Parizotto EA, Himber C, Voinnet O. Moissiard G, et al. RNA. 2007 Aug;13(8):1268-78. doi: 10.1261/rna.541307. Epub 2007 Jun 25. RNA. 2007. PMID: 17592042 Free PMC article. - A novel class of bacteria-induced small RNAs in Arabidopsis.
Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H. Katiyar-Agarwal S, et al. Genes Dev. 2007 Dec 1;21(23):3123-34. doi: 10.1101/gad.1595107. Epub 2007 Nov 14. Genes Dev. 2007. PMID: 18003861 Free PMC article. - siRNA and miRNA processing: new functions for Cajal bodies.
Pontes O, Pikaard CS. Pontes O, et al. Curr Opin Genet Dev. 2008 Apr;18(2):197-203. doi: 10.1016/j.gde.2008.01.008. Epub 2008 Mar 11. Curr Opin Genet Dev. 2008. PMID: 18337083 Free PMC article. Review. - Endogenous small interfering RNAs in animals.
Okamura K, Lai EC. Okamura K, et al. Nat Rev Mol Cell Biol. 2008 Sep;9(9):673-8. doi: 10.1038/nrm2479. Nat Rev Mol Cell Biol. 2008. PMID: 18719707 Free PMC article. Review.
Cited by
- Small RNA profiling reveals phosphorus deficiency as a contributing factor in symptom expression for citrus huanglongbing disease.
Zhao H, Sun R, Albrecht U, Padmanabhan C, Wang A, Coffey MD, Girke T, Wang Z, Close TJ, Roose M, Yokomi RK, Folimonova S, Vidalakis G, Rouse R, Bowman KD, Jin H. Zhao H, et al. Mol Plant. 2013 Mar;6(2):301-10. doi: 10.1093/mp/sst002. Epub 2013 Jan 5. Mol Plant. 2013. PMID: 23292880 Free PMC article. - Novel Insights into Insect-Microbe Interactions-Role of Epigenomics and Small RNAs.
Kim D, Thairu MW, Hansen AK. Kim D, et al. Front Plant Sci. 2016 Aug 4;7:1164. doi: 10.3389/fpls.2016.01164. eCollection 2016. Front Plant Sci. 2016. PMID: 27540386 Free PMC article. Review. - Long small RNA76113 targets CYCLIC NUCLEOTIDE-GATED ION CHANNEL 5 to repress disease resistance in rice.
Zheng L, Yu Y, Zheng Y, Wang Y, Wu N, Jiang C, Zhao H, Niu D. Zheng L, et al. Plant Physiol. 2024 Feb 29;194(3):1889-1905. doi: 10.1093/plphys/kiad599. Plant Physiol. 2024. PMID: 37949839 Free PMC article. - High throughput sequencing reveals novel and abiotic stress-regulated microRNAs in the inflorescences of rice.
Barrera-Figueroa BE, Gao L, Wu Z, Zhou X, Zhu J, Jin H, Liu R, Zhu JK. Barrera-Figueroa BE, et al. BMC Plant Biol. 2012 Aug 3;12:132. doi: 10.1186/1471-2229-12-132. BMC Plant Biol. 2012. PMID: 22862743 Free PMC article. - Identification of miRNAs and Their Targets in Cotton Inoculated with Verticillium dahliae by High-Throughput Sequencing and Degradome Analysis.
Zhang Y, Wang W, Chen J, Liu J, Xia M, Shen F. Zhang Y, et al. Int J Mol Sci. 2015 Jun 30;16(7):14749-68. doi: 10.3390/ijms160714749. Int J Mol Sci. 2015. PMID: 26133244 Free PMC article.
References
- Baulcombe D. Trends Biochem Sci. 2005;30:290–293. - PubMed
- Sontheimer EJ, Carthew RW. Cell. 2005;122:9–12. - PubMed
- Jones-Rhoades M, Bartel D, Bartel B. Annu Rev Plant Biol. 2006;57:19–53. - PubMed
- Mallory AC, Vaucheret H. Nat Genet. 2006;38:S31–S36. - PubMed
- Jones-Rhoades MW, Bartel DP. Mol Cell. 2004;14:787–799. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM070795/GM/NIGMS NIH HHS/United States
- R01-FM069680-01/PHS HHS/United States
- R01 GM059138/GM/NIGMS NIH HHS/United States
- R01GM59138/GM/NIGMS NIH HHS/United States
- R01GM070795/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous