The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases - PubMed (original) (raw)
The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases
Tohru Ono et al. Proc Natl Acad Sci U S A. 2006.
Abstract
The silent information regulator 2 (Sir2) family of NAD-dependent N-acetyl-protein deacetylases participates in the regulation of gene silencing, chromatin structure, and longevity. In the Sir2-catalyzed reaction, the acetyl moiety of N-acetyl-histone is transferred to the ADP-ribose of NAD, yielding O-acetyl-ADP-ribose and nicotinamide. We hypothesized that, if O-acetyl-ADP-ribose were an important signaling molecule, a specific hydrolase would cleave the (O-acetyl)-(ADP-ribose) linkage. We report here that the poly(ADP-ribose) glycohydrolase ARH3 hydrolyzed O-acetyl-ADP-ribose to produce ADP-ribose in a time- and Mg(2+)-dependent reaction and thus could participate in two signaling pathways. This O-acetyl-ADP-ribose hydrolase belongs to a family of three structurally related 39-kDa ADP-ribose-binding proteins (ARH1-ARH3). ARH1 was reported to hydrolyze ADP-ribosylarginine, whereas ARH3 degraded poly(ADP-ribose). ARH3-catalyzed generation of ADP-ribose from O-acetyl-ADP-ribose was significantly faster than from poly(ADP-ribose). Like the degradation of poly(ADP-ribose) by ARH3, hydrolysis of O-acetyl-ADP-ribose was abolished by replacement of the vicinal aspartates at positions 77 and 78 of ARH3 with asparagine. The rate of O-acetyl-ADP-ribose hydrolysis by recombinant ARH3 was 250-fold that observed with ARH1; ARH2 and poly(ADP-ribose) glycohydrolase were inactive. All data support the conclusion that the Sir2 reaction product O-acetyl-ADP-ribose is degraded by ARH3.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
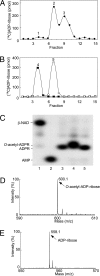
Identification of products of SIRT1- and ARH3-catalyzed reactions. (A) Synthesis of _O-_acetyl-[14C]ADP-ribose catalyzed by SIRT1. Data are expressed as picomoles of 14C per fraction. Peaks: 1, ADP-ribose; 2, _O-_acetyl-ADP-ribose; 3, β-NAD. The results of duplicate assays in three experiments were averaged (n = 6; average ± SD.). (B) Hydrolysis of _O-_acetyl-[14C]ADP-ribose catalyzed by ARH3. Peaks: 4, ADP-ribose; 5, _O-_acetyl-ADP-ribose. The results of duplicate assays in three experiments were averaged (n = 6; average ± SD). (C) High-resolution polyacrylamide gel electrophoresis of substrates and products in reactions involving of _O-_acetyl-[32P]ADP-ribose. Lane 1, [32P]NAD. Lane 2, [32P]AMP produced by pyrophosphatase cleavage of [32P]NAD (see Materials and Methods for details). Lane 3, [32P]ADP-ribose produced from [32P]NAD by CTA glycohydrolase activity (see Materials and Methods for details). Lane 4, _O-_acetyl-[32P]ADP-ribose synthesized by SIRT1 as in Materials and Methods. Lane 5, [32P]ADP-ribose produced by ARH3 from _O-_acetyl-[32P]ADP-ribose as in Materials and Methods. All assays were run in duplicate. Results were similar in three experiments. (D) Identification of _O-_acetyl-ADP-ribose by MALDI-TOF mass spectrometry analysis (see Materials and Methods for details). MALDI-TOF mass spectrometry was used to identify the RP-HPLC products; a molecule of 600 m/z was identified, consistent with the formation of _O-_acetyl-ADP-ribose. (E) Identification of the reaction products formed by ARH3 from _O-_acetyl-ADP-ribose. We identified a product of 558 m/z, consistent with the predicted mass of ADP-ribose.
Fig. 2.
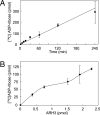
Hydrolysis of _O-_acetyl-ADP-ribose by ARH3. (A) Hydrolysis of _O-_acetyl-ADP-ribose by ARH3. Assays containing 1.5 pmol of mouse ARH3 and 2.5 μM _O-_acetyl-[14C]ADP-ribose, 50 mM potassium phosphate (pH 7.0), 10 mM MgCl2, and 5 mM DTT (total volume 200 μl) were incubated at 30°C for the indicated time before separation of substrate and products by using RP-HPLC as described in Materials and Methods. Data are means ± 1/2 the range of values from duplicate assays. Results were similar in two experiments. (B) Assays containing 2.5 μM _O-_acetyl-[14C]ADP-ribose and the indicated amount of mouse ARH3 in 200 μl of buffer were incubated for 1 h at 30°C before separation of substrate and products by using RP-HPLC as described in Materials and Methods. Data are means ± 1/2 the range of values from duplicate assays. Results were similar in two experiments.
Fig. 3.
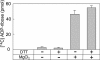
Effect of DTT and Mg2+ on _O-_acetyl-ADP-ribose hydrolase activity. Assays with 1.5 pmol of mouse ARH3 and 2.5 μM _O-_acetyl-[14C]ADP-ribose, 50 mM potassium phosphate (pH 7.0), and with or without 5 mM DTT and/or 10 mM MgCl2 (total volume 200 μl) were incubated for 1 h at 30°C before separation of substrate and products by using RP-HPLC as described in Materials and Methods. Data are means ± 1/2 the range of values from duplicate assays. Results were similar in two experiments.
Fig. 4.
Hydrolysis of _O-_acetyl-ADP-ribose by ARH1, ARH2, and ARH3. (A) Hydrolysis of _O-_acetyl-[14C]ADP-ribose by ARH1, ARH2, and ARH3. Assays with the indicated amount of mouse ARH1(●), ARH2 (■), and ARH3 (○) and 2.5 μM _O-_acetyl-[14C]ADP-ribose, 50 mM potassium phosphate (pH 7.0), 10 mM MgCl2, and 5 mM DTT (total volume, 200 μl) were incubated for 2 h at 30°C before separation of substrate and products by using RP-HPLC as described in Materials and Methods. Data are means ± 1/2 the range of values from duplicate assays. Results were similar in two experiments. (B) Assays containing 230 pmol of mouse ARH1 in 200 μl of buffer were incubated at 30°C for the indicated time before separation of substrate and products by using RP-HPLC as described in Materials and Methods. Data are means ± 1/2 the range of values from duplicate assays. Results were similar in two experiments.
Fig. 5.
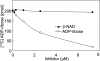
Inhibition assay of ARH3 hydrolase activity by ADP-ribose and β-NAD. Assays containing 2 pmol of mouse ARH3 and 2.5 μM _O-_acetyl-[14C]ADP-ribose and the indicated amount of β-NAD (●) or ADP-ribose (○), 50 mM potassium phosphate (pH 7.0), 10 mM MgCl2, and 5 mM DTT (total volume 200 μl) were incubated for 2 h at 30°C before separation of substrate and products by using RP-HPLC as described in Materials and Methods. Data are means ± 1/2 the range of values from duplicate assays. Results were similar in three experiments.
Fig. 6.
Hydrolysis of _O-_acetyl-ADP-ribose by wild-type and mutant forms of ARH3 or PARG. Assays with 1.5 pmol of wild-type or mutant human (D77/78N) ARH3 or 20 pmol of PARG and 2.5 μM _O-_acetyl-[14C]ADP-ribose, 50 mM potassium phosphate (pH 7.0), 10 mM MgCl2, and 5 mM DTT (total volume, 200 μl) were incubated for 2 h at 30°C before separation of substrate and products by using RP-HPLC as described in Materials and Methods. All assays were run in duplicate. Data are means ± 1/2 the range of values from duplicate assays. Results were similar in two experiments. *D77/78N, assays incubated with 15 pmol of mutant human ARH3 (D77/78N).
Similar articles
- Hydrolysis of O-acetyl-ADP-ribose isomers by ADP-ribosylhydrolase 3.
Kasamatsu A, Nakao M, Smith BC, Comstock LR, Ono T, Kato J, Denu JM, Moss J. Kasamatsu A, et al. J Biol Chem. 2011 Jun 17;286(24):21110-7. doi: 10.1074/jbc.M111.237636. Epub 2011 Apr 17. J Biol Chem. 2011. PMID: 21498885 Free PMC article. - Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase.
Oka S, Kato J, Moss J. Oka S, et al. J Biol Chem. 2006 Jan 13;281(2):705-13. doi: 10.1074/jbc.M510290200. Epub 2005 Nov 8. J Biol Chem. 2006. PMID: 16278211 - ARH Family of ADP-Ribose-Acceptor Hydrolases.
Ishiwata-Endo H, Kato J, Yamashita S, Chea C, Koike K, Lee DY, Moss J. Ishiwata-Endo H, et al. Cells. 2022 Nov 30;11(23):3853. doi: 10.3390/cells11233853. Cells. 2022. PMID: 36497109 Free PMC article. Review. - ADP-Ribosyl-Acceptor Hydrolase Activities Catalyzed by the ARH Family of Proteins.
Mashimo M, Moss J. Mashimo M, et al. Methods Mol Biol. 2018;1813:187-204. doi: 10.1007/978-1-4939-8588-3_12. Methods Mol Biol. 2018. PMID: 30097868 - Structure and function of the ARH family of ADP-ribosyl-acceptor hydrolases.
Mashimo M, Kato J, Moss J. Mashimo M, et al. DNA Repair (Amst). 2014 Nov;23:88-94. doi: 10.1016/j.dnarep.2014.03.005. Epub 2014 Apr 18. DNA Repair (Amst). 2014. PMID: 24746921 Free PMC article. Review.
Cited by
- Acylation of Biomolecules in Prokaryotes: a Widespread Strategy for the Control of Biological Function and Metabolic Stress.
Hentchel KL, Escalante-Semerena JC. Hentchel KL, et al. Microbiol Mol Biol Rev. 2015 Sep;79(3):321-46. doi: 10.1128/MMBR.00020-15. Epub 2015 Jul 15. Microbiol Mol Biol Rev. 2015. PMID: 26179745 Free PMC article. Review. - Mitochondrial dysfunction and NAD(+) metabolism alterations in the pathophysiology of acute brain injury.
Owens K, Park JH, Schuh R, Kristian T. Owens K, et al. Transl Stroke Res. 2013 Dec;4(6):618-34. doi: 10.1007/s12975-013-0278-x. Epub 2013 Aug 10. Transl Stroke Res. 2013. PMID: 24323416 Review. - Mechanistic insights into the three steps of poly(ADP-ribosylation) reversal.
Rack JGM, Liu Q, Zorzini V, Voorneveld J, Ariza A, Honarmand Ebrahimi K, Reber JM, Krassnig SC, Ahel D, van der Marel GA, Mangerich A, McCullagh JSO, Filippov DV, Ahel I. Rack JGM, et al. Nat Commun. 2021 Jul 28;12(1):4581. doi: 10.1038/s41467-021-24723-3. Nat Commun. 2021. PMID: 34321462 Free PMC article. - Poly-ADP ribosylation in DNA damage response and cancer therapy.
Hou WH, Chen SH, Yu X. Hou WH, et al. Mutat Res Rev Mutat Res. 2019 Apr-Jun;780:82-91. doi: 10.1016/j.mrrev.2017.09.004. Epub 2017 Sep 20. Mutat Res Rev Mutat Res. 2019. PMID: 31395352 Free PMC article. Review. - The natural history of ADP-ribosyltransferases and the ADP-ribosylation system.
Aravind L, Zhang D, de Souza RF, Anand S, Iyer LM. Aravind L, et al. Curr Top Microbiol Immunol. 2015;384:3-32. doi: 10.1007/82_2014_414. Curr Top Microbiol Immunol. 2015. PMID: 25027823 Free PMC article. Review.
References
- Guarente L. Genes Dev. 2000;14:1021–1026. - PubMed
- Borra MT, O'Neill FJ, Jackson MD, Marshall B, Verdin E, Foltz KR, Denu JM. J Biol Chem. 2002;277:12632–12641. - PubMed
- Jackson MD, Denu JM. J Biol Chem. 2002;277:18535–18544. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases