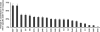HLA Alleles Associated with Delayed Progression to AIDS Contribute Strongly to the Initial CD8(+) T Cell Response against HIV-1 - PubMed (original) (raw)
Multicenter Study
doi: 10.1371/journal.pmed.0030403.
Elizabeth T Kalife, Ying Qi, Hendrik Streeck, Mathias Lichterfeld, Mary N Johnston, Nicole Burgett, Martha E Swartz, Amy Yang, Galit Alter, Xu G Yu, Angela Meier, Juergen K Rockstroh, Todd M Allen, Heiko Jessen, Eric S Rosenberg, Mary Carrington, Bruce D Walker
- PMID: 17076553
- PMCID: PMC1626551
- DOI: 10.1371/journal.pmed.0030403
Multicenter Study
HLA Alleles Associated with Delayed Progression to AIDS Contribute Strongly to the Initial CD8(+) T Cell Response against HIV-1
Marcus Altfeld et al. PLoS Med. 2006 Oct.
Abstract
Background: Very little is known about the immunodominance patterns of HIV-1-specific T cell responses during primary HIV-1 infection and the reasons for human lymphocyte antigen (HLA) modulation of disease progression.
Methods and findings: In a cohort of 104 individuals with primary HIV-1 infection, we demonstrate that a subset of CD8(+) T cell epitopes within HIV-1 are consistently targeted early after infection, while other epitopes subsequently targeted through the same HLA class I alleles are rarely recognized. Certain HLA alleles consistently contributed more than others to the total virus-specific CD8(+) T cell response during primary infection, and also reduced the absolute magnitude of responses restricted by other alleles if coexpressed in the same individual, consistent with immunodomination. Furthermore, individual HLA class I alleles that have been associated with slower HIV-1 disease progression contributed strongly to the total HIV-1-specific CD8(+) T cell response during primary infection.
Conclusions: These data demonstrate consistent immunodominance patterns of HIV-1-specific CD8(+) T cell responses during primary infection and provide a mechanistic explanation for the protective effect of specific HLA class I alleles on HIV-1 disease progression.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1
Immunodominance Patterns for HIV-1-Specific CD8+ T Cell Responses Restricted by Individual HLA Class I Alleles Peptides corresponding to described optimal HIV-1-specific CD8+ T cell epitopes were tested in all study participants expressing the respective HLA class I allele. The average magnitude of CD8+ T cell responses specific for each tested peptide, given as SFCs per million input PBMCs (SFC/Mill PBMC), are shown as bars in the left part of each graph. The percentage of participants expressing the respective allele that had a detectable peptide-specific CD8+ T cell response are shown as bars on the right part of each graph. Epitopes are aligned for each HLA class I allele according to their frequency of recognition from top to bottom. The peptide number corresponds to the peptide sequence listed for each HLA allele and number in Table 2. Data are shown only for HLA class I alleles that were expressed in at least three individuals, and for which at least three HIV-1-specific optimal CD8+ T cell epitopes had been defined.
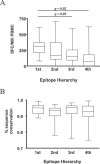
Figure 2. CD8+ T Cell responses Directed against the Most Frequently Recognized CD8+ T Cell Epitopes Were Also of the Highest Magnitude
(A) For each HLA class I allele studied, the four most frequently targeted HIV-1-specific CD8+ T cell epitopes were listed according to their hierarchy (for HLA class I alleles with only three described epitopes, the fourth value was excluded from the analysis). The average magnitudes of CD8+ T cell responses directed against the 1st, 2nd, 3rd, and 4th most frequently targeted epitope for each allele were calculated (given as SFCs per million input PBMCs [SFC/Mill PBMC]) and are shown as box plots. The average magnitude of the most frequently targeted HIV-1-specific CD8+ T cell epitopes restricted by each allele were significantly higher than the average magnitude of the 3rd and 4th most frequently targeted epitopes, and the respective _p_-values are provided above the box plot. (B) The sequence conservation within targeted CD8+ T cell epitopes does not contribute significantly to the observed immunodominance patterns of HIV-1-specific CD8+ T cell response in primary infection. For each HLA class I alleles studied, the four most frequently targeted HIV-1-specific CD8+ T cell epitopes were listed according to their hierarchy (for HLA class I alleles with only three described epitopes; the fourth value was excluded from the analysis). The average sequence conservations, in comparison to HIV-1 clade B sequences published in the Los Alamos Database, of the 1st, 2nd, 3rd, and 4th most frequently targeted epitope for each allele were calculated (given as percent sequence conservation) and are shown as box plots. The average percentage of sequence conservation of the four most frequently targeted HIV-1-specific CD8+ T cell epitopes restricted by each allele did not differ significantly.
Figure 3. Different HLA Class I Alleles Differ in Their Contribution to the Total HIV-1-Specific CD8+ T Cell Response
The percentage contribution of HIV-1-specific CD8+ T cell responses restricted by each individual HLA class I allele to the total HIV-1-specific CD8+ T cell response in individuals expressing the respective allele is shown. HLA class I alleles are listed according to their contribution from left to right. HLA-B57 and HLA-B27 contributed 66% and 65.4%, respectively, to the total HIV-1-specific CD8+ T cell response in individuals expressing these alleles.
Figure 4. Immunodomination of HLA-B57- and HLA-B27-Restricted HIV-1-Specific CD8+ T Cell Responses
The percent contribution (left graphs) and the absolute magnitude (right graphs, given as SFCs per million input PBMCs [SFC/Mill PBMC]) of HLA-A1-, -A2-, -A3-, and -A24-restricted HIV-1-specific CD8+ T cell responses in individuals expressing these HLA class I alleles alone, or in conjunction with HLA-B57 or HLA-B27, are shown. Each dot represents data for one individual. The contribution, as well as the absolute magnitude, of HIV-1-specific CD8+ T cell responses directed against HLA-A1-, -A2-, and -A24-restricted CD8+ T cell epitopes was significantly lower in participants that also coexpressed HLA-B57 or HLA-B27. The same trend was observed for HLA-A3, but did not reach statistical significance. HLA-A1-, -A2-, -A3-, and -A24-restricted HIV-1-specific CD8+ T cell response did not differ between individuals expressing other frequent HLA class B alleles, such as HLA-B7, -B8, -B35, or -B44 (unpublished data).
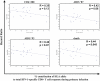
Figure 5. Correlation between the Contribution of Individual HLA Class I Alleles to the Total HIV-1-Specific CD8+ T Cell Response during Primary Infection and the HR for HIV-1 Infection Outcome
The percent contribution of individual HLA class I alleles to the total HIV-1-specific CD8+ T cell response during primary infection was correlated to the HR for four different HIV-1 infection outcomes (time to CD4 <200, time to AIDS 1987, time to AIDS 1993, and time to death) for the respective HLA alleles.
Similar articles
- HIV-1-specific cytotoxic T lymphocyte (CTL) responses against immunodominant optimal epitopes slow the progression of AIDS in China.
Zhai S, Zhuang Y, Song Y, Li S, Huang D, Kang W, Li X, Liao Q, Liu Y, Zhao Z, Lu Y, Sun Y. Zhai S, et al. Curr HIV Res. 2008 Jun;6(4):335-50. doi: 10.2174/157016208785132473. Curr HIV Res. 2008. PMID: 18691032 - Consistent patterns in the development and immunodominance of human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T-cell responses following acute HIV-1 infection.
Yu XG, Addo MM, Rosenberg ES, Rodriguez WR, Lee PK, Fitzpatrick CA, Johnston MN, Strick D, Goulder PJ, Walker BD, Altfeld M. Yu XG, et al. J Virol. 2002 Sep;76(17):8690-701. doi: 10.1128/jvi.76.17.8690-8701.2002. J Virol. 2002. PMID: 12163589 Free PMC article. - Differential Immunodominance Hierarchy of CD8+ T-Cell Responses in HLA-B*27:05- and -B*27:02-Mediated Control of HIV-1 Infection.
Adland E, Hill M, Lavandier N, Csala A, Edwards A, Chen F, Radkowski M, Kowalska JD, Paraskevis D, Hatzakis A, Valenzuela-Ponce H, Pfafferott K, Williams I, Pellegrino P, Borrow P, Mori M, Rockstroh J, Prado JG, Mothe B, Dalmau J, Martinez-Picado J, Tudor-Williams G, Frater J, Stryhn A, Buus S, Teran GR, Mallal S, John M, Buchbinder S, Kirk G, Martin J, Michael N, Fellay J, Deeks S, Walker B, Avila-Rios S, Cole D, Brander C, Carrington M, Goulder P. Adland E, et al. J Virol. 2018 Jan 30;92(4):e01685-17. doi: 10.1128/JVI.01685-17. Print 2018 Feb 15. J Virol. 2018. PMID: 29167337 Free PMC article. - [Protective role of HLA-B27 in HIV and hepatitis C virus infection].
Neumann-Haefelin C. Neumann-Haefelin C. Dtsch Med Wochenschr. 2011 Feb;136(7):320-4. doi: 10.1055/s-0031-1272531. Epub 2011 Feb 7. Dtsch Med Wochenschr. 2011. PMID: 21302207 Review. German. - Progression of HIV to AIDS: a protective role for HLA-B27?
den Uyl D, van der Horst-Bruinsma IE, van Agtmael M. den Uyl D, et al. AIDS Rev. 2004 Apr-Jun;6(2):89-96. AIDS Rev. 2004. PMID: 15332431 Review.
Cited by
- Modelling the spread of HIV immune escape mutants in a vaccinated population.
Fryer HR, McLean AR. Fryer HR, et al. PLoS Comput Biol. 2011 Dec;7(12):e1002289. doi: 10.1371/journal.pcbi.1002289. Epub 2011 Dec 1. PLoS Comput Biol. 2011. PMID: 22144883 Free PMC article. - Variable processing and cross-presentation of HIV by dendritic cells and macrophages shapes CTL immunodominance and immune escape.
Dinter J, Duong E, Lai NY, Berberich MJ, Kourjian G, Bracho-Sanchez E, Chu D, Su H, Zhang SC, Le Gall S. Dinter J, et al. PLoS Pathog. 2015 Mar 17;11(3):e1004725. doi: 10.1371/journal.ppat.1004725. eCollection 2015 Mar. PLoS Pathog. 2015. PMID: 25781895 Free PMC article. - Characterization of a proteasome and TAP-independent presentation of intracellular epitopes by HLA-B27 molecules.
Magnacca A, Persiconi I, Nurzia E, Caristi S, Meloni F, Barnaba V, Paladini F, Raimondo D, Fiorillo MT, Sorrentino R. Magnacca A, et al. J Biol Chem. 2012 Aug 31;287(36):30358-67. doi: 10.1074/jbc.M112.384339. Epub 2012 Jul 17. J Biol Chem. 2012. PMID: 22807446 Free PMC article. - Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover.
Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D, Bornstein E, Asher TE, Samri A, Schnuriger A, Theodorou I, Costagliola D, Rouzioux C, Agut H, Marcelin AG, Douek D, Autran B, Appay V. Almeida JR, et al. J Exp Med. 2007 Oct 1;204(10):2473-85. doi: 10.1084/jem.20070784. Epub 2007 Sep 24. J Exp Med. 2007. PMID: 17893201 Free PMC article. - Prolonged control of an HIV type 1 escape variant following treatment interruption in an HLA-B*27-positive patient.
O'Connell KA, Pelz RK, Dinoso JB, Dunlop E, Paik-Tesch J, Williams TM, Blankson JN. O'Connell KA, et al. AIDS Res Hum Retroviruses. 2010 Dec;26(12):1307-11. doi: 10.1089/aid.2010.0135. Epub 2010 Sep 21. AIDS Res Hum Retroviruses. 2010. PMID: 20854198 Free PMC article.
References
- Kahn JO, Walker BD. Acute human immunodeficiency virus type 1 infection. N Engl J Med. 1998;339:33–39. - PubMed
- Hecht FM, Busch MP, Rawal B, Webb M, Rosenberg E, et al. Use of laboratory tests and clinical symptoms for identification of primary HIV infection. AIDS. 2002;16:1119–1129. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials