The tree of one percent - PubMed (original) (raw)
The tree of one percent
Tal Dagan et al. Genome Biol. 2006.
Abstract
Two significant evolutionary processes are fundamentally not tree-like in nature--lateral gene transfer among prokaryotes and endosymbiotic gene transfer (from organelles) among eukaryotes. To incorporate such processes into the bigger picture of early evolution, biologists need to depart from the preconceived notion that all genomes are related by a single bifurcating tree.
Figures
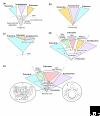
Figure 1
Five different current views of the general shape of microbial evolution. (a) The 'classical' tree derived from comparison of rRNA sequence and rooted with ancient paralogs. It is thought to arise from a collection of non-cellular supramolecular aggregates in the primordial soup, between which there is lateral gene transfer (LGT). A process dubbed genetic annealing gives rise to cells. In this scenario, the three domains of life - Eubacteria, Archaebacteria and Eukaryotes - branch off in that order. (b) The introns-early tree. This proposes that the ancestor of all three domains contained introns, which were lost in the Archaebacteria and Euacteria. (c) The neomuran tree. This introduces an ancestral group of organisms from which Archaeabacteria and Eukaryotes arose after the loss of the eubacterial-type cell wall in one lineage (the neomuran revolution). (d) The symbiotic tree. This proposes that the ancestor of eukaryotes originated by the endosymbiosis of one prokaryote (X) in another prokaryote host (Y), giving rise to nucleated (n) eukaryotic cells. The different groups of eukaryotes arose by subsequent separate endosymbiotic events involving various prokaryotes - the ancestors of plastids (p) and mitochondria (m) - in host cells of this lineage. (e) The prokaryote-host tree. This also incorporates endosymbiosis as the origin of mitochondria and plastids, but proposes that the endosymbiotic event that gave rise to a cell containing nucleus and mitochondria occurred in a prokaryotic host. This leads to a ring-like relationship between the ancestral organisms rather than a tree (see inset 2). This model also invokes extensive LGT throughout microbial evolution (see inset 1). See text for further details.
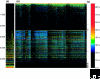
Figure 2
As a representative eukaryote example, the non-redundant set of human proteins (NCBI's Refseq database [70]) was compared using BLAST to a data set containing all proteins from 224 prokaryotic genomes: (a) 24 archaebacteria and (b) 200 eubacteria. In each panel, individual genomes are represented by columns and individual proteins by rows; numbers of proteins are indicated on the left and percentage amino-acid identity by the color scale shown on the right. BLAST hits with an e-value ≤ 10-20 and ≥ 20% amino-acid identity were recorded. The percent identity of the best blast hit for each human protein in each prokaryote was color coded as shown on the right and plotted with MATLAB©. The 31 proteins that were used in the recent tree of life [9] are marked with ticks in column (c). A table containing the numbers, genes, and species underlying the figure is available as additional data file 1.
Similar articles
- Getting a better picture of microbial evolution en route to a network of genomes.
Dagan T, Martin W. Dagan T, et al. Philos Trans R Soc Lond B Biol Sci. 2009 Aug 12;364(1527):2187-96. doi: 10.1098/rstb.2009.0040. Philos Trans R Soc Lond B Biol Sci. 2009. PMID: 19571239 Free PMC article. Review. - The tree and net components of prokaryote evolution.
Puigbò P, Wolf YI, Koonin EV. Puigbò P, et al. Genome Biol Evol. 2010;2:745-56. doi: 10.1093/gbe/evq062. Epub 2010 Oct 1. Genome Biol Evol. 2010. PMID: 20889655 Free PMC article. - Origin of the bacterial SET domain genes: vertical or horizontal?
Alvarez-Venegas R, Sadder M, Tikhonov A, Avramova Z. Alvarez-Venegas R, et al. Mol Biol Evol. 2007 Feb;24(2):482-97. doi: 10.1093/molbev/msl184. Epub 2006 Dec 5. Mol Biol Evol. 2007. PMID: 17148507 - Genomes in flux: the evolution of archaeal and proteobacterial gene content.
Snel B, Bork P, Huynen MA. Snel B, et al. Genome Res. 2002 Jan;12(1):17-25. doi: 10.1101/gr.176501. Genome Res. 2002. PMID: 11779827 - Phylogenomic networks.
Dagan T. Dagan T. Trends Microbiol. 2011 Oct;19(10):483-91. doi: 10.1016/j.tim.2011.07.001. Epub 2011 Aug 3. Trends Microbiol. 2011. PMID: 21820313 Review.
Cited by
- What Is Speciation?
Shapiro BJ, Leducq JB, Mallet J. Shapiro BJ, et al. PLoS Genet. 2016 Mar 31;12(3):e1005860. doi: 10.1371/journal.pgen.1005860. eCollection 2016 Mar. PLoS Genet. 2016. PMID: 27030977 Free PMC article. Review. - The last universal common ancestor between ancient Earth chemistry and the onset of genetics.
Weiss MC, Preiner M, Xavier JC, Zimorski V, Martin WF. Weiss MC, et al. PLoS Genet. 2018 Aug 16;14(8):e1007518. doi: 10.1371/journal.pgen.1007518. eCollection 2018 Aug. PLoS Genet. 2018. PMID: 30114187 Free PMC article. Review. - Prokaryotic evolution and the tree of life are two different things.
Bapteste E, O'Malley MA, Beiko RG, Ereshefsky M, Gogarten JP, Franklin-Hall L, Lapointe FJ, Dupré J, Dagan T, Boucher Y, Martin W. Bapteste E, et al. Biol Direct. 2009 Sep 29;4:34. doi: 10.1186/1745-6150-4-34. Biol Direct. 2009. PMID: 19788731 Free PMC article. - Statistics and truth in phylogenomics.
Kumar S, Filipski AJ, Battistuzzi FU, Kosakovsky Pond SL, Tamura K. Kumar S, et al. Mol Biol Evol. 2012 Feb;29(2):457-72. doi: 10.1093/molbev/msr202. Epub 2011 Aug 26. Mol Biol Evol. 2012. PMID: 21873298 Free PMC article. Review. - The tree versus the forest: the fungal tree of life and the topological diversity within the yeast phylome.
Marcet-Houben M, Gabaldón T. Marcet-Houben M, et al. PLoS One. 2009;4(2):e4357. doi: 10.1371/journal.pone.0004357. Epub 2009 Feb 3. PLoS One. 2009. PMID: 19190756 Free PMC article.
References
- Doolittle WF. If the tree of life fell, would it make a sound? In: Sapp J, editor. Microbial Phylogeny and Evolution: Concepts and Controversies. New York: Oxford University Press; 2004. pp. 119–133.
- Mirkin BG, Fenner TI, Galperin MY, Koonin EV. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol Biol. 2003;3:2. doi: 10.1186/1471-2148-3-2. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources