Genomic analysis of pilocytic astrocytomas at 0.97 Mb resolution shows an increasing tendency toward chromosomal copy number change with age - PubMed (original) (raw)
Genomic analysis of pilocytic astrocytomas at 0.97 Mb resolution shows an increasing tendency toward chromosomal copy number change with age
David T W Jones et al. J Neuropathol Exp Neurol. 2006 Nov.
Abstract
Brain tumors are the most common solid tumors of childhood, accounting for over 20% of cancers in children under 15 years of age. Pilocytic astrocytomas (PAs), World Health Organization grade I, are one of the most frequently occurring childhood brain tumors, yet little is known about genetic changes characterizing this entity. We have used microarray comparative genomic hybridization at 0.97 Mb resolution to study a series of PAs (n = 44). No copy number abnormality was seen in 64% of cases at this resolution. However, whole chromosomal gain (median 5 chromosomes affected) occurred in 32% of tumors. The most frequently affected chromosomes were 5 and 7 (11 of 44 cases each) followed by 6, 11, 15, and 20 (greater than 10% of cases each). Findings were confirmed by fluorescence in situ hybridization and microsatellite analysis in a subset of tumors. Chromosomal gain was significantly more frequent in PAs from patients over 15 years old (p = 0.03, Fisher exact test). The number of chromosomes involved was also significantly greater in the older group (p = 0.02, Mann-Whitney U test). One case (2%) showed a region of gain on chromosome 3 and one (2%) a deletion on 6q as their sole abnormalities. This is the first genomewide study to show this nonrandom pattern of genetic alteration in pilocytic astrocytomas.
Figures
FIGURE 1
Whole genome microarray comparative genomic hybridization (aCGH) plot of DNA copy number ratio for sample PA29 showing gain of chromosomes 5, 6, 7, 8, and 11. Insets show microsatellite data from markers in chromosomes 1, 6, 7, and 11. Note the increase in the intensity of the tumor alleles indicating a gain of genetic material on chromosomes 6, 7, and 11 correlating with the gains shown in the aCGH plot. Microsatellite analysis of chromosome 1 indicates balance, again correlating with the aCGH data.
FIGURE 2
Part of a whole genome microarray comparative genomic hybridization plot from sample PA20 showing DNA copy number ratio across chromosome 3, including a region of gain on 3p.
FIGURE 3
Part of a whole genome microarray comparative genomic hybridization plot from sample PA47 showing DNA copy number ratio across chromosome 6. The inset shows the region from a chromosome 6 tiling path array corresponding to the region of loss at 6q26 seen on the 0.97 Mb resolution plot.
FIGURE 4
DNA copy number ratio across chromosome 3 for PA35 indicating a pericentromeric region of gain.
FIGURE 5
Summary of changes found in 44 primary tumor samples grouped to show similar patterns of chromosomal gain. Cases with no chromosomal gain are in descending age order. The age of each case is shown with the case number (n/a, not available). Adult cases are shown on the top and childhood cases underneath with chromosome number indicated below both. Each filled box represents a chromosomal gain. *, Small region of gain; ×, small region of loss.
FIGURE 6
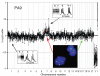
A whole genome plot for PA9 showing a subtle gain of chromosome 7 confirmed by the gain of intensity of the tumor allele seen on the inset. A fluorescent in situ hybridization image of a section from PA9 is also shown. Chr7 centromeric probes are shown in red and Chr17 centromeric probes in green with a blue DAPI counterstain. Positions of centromeres of chromosomes 7 and 17 on the microarray comparative genomic hybridization plot are shown with a red and green dot, respectively. After immunohistochemical analysis with CD68k and CD45, this case had a revised estimate of tumor cell content of only 66%.
Similar articles
- Frequent gains at chromosome 7q34 involving BRAF in pilocytic astrocytoma.
Bar EE, Lin A, Tihan T, Burger PC, Eberhart CG. Bar EE, et al. J Neuropathol Exp Neurol. 2008 Sep;67(9):878-87. doi: 10.1097/NEN.0b013e3181845622. J Neuropathol Exp Neurol. 2008. PMID: 18716556 - Nonrandom chromosomal gains in pilocytic astrocytomas of childhood.
White FV, Anthony DC, Yunis EJ, Tarbell NJ, Scott RM, Schofield DE. White FV, et al. Hum Pathol. 1995 Sep;26(9):979-86. doi: 10.1016/0046-8177(95)90087-x. Hum Pathol. 1995. PMID: 7672798 - Are juvenile pilocytic astrocytomas benign tumors? A cytogenetic study in 24 cases.
Zattara-Cannoni H, Gambarelli D, Lena G, Dufour H, Choux M, Grisoli F, Vagner-Capodano AM. Zattara-Cannoni H, et al. Cancer Genet Cytogenet. 1998 Jul 15;104(2):157-60. doi: 10.1016/s0165-4608(97)00455-x. Cancer Genet Cytogenet. 1998. PMID: 9666811 Review. - Investigation of genetic alterations associated with the grade of astrocytic tumor by comparative genomic hybridization.
Nishizaki T, Ozaki S, Harada K, Ito H, Arai H, Beppu T, Sasaki K. Nishizaki T, et al. Genes Chromosomes Cancer. 1998 Apr;21(4):340-6. doi: 10.1002/(sici)1098-2264(199804)21:4<340::aid-gcc8>3.0.co;2-z. Genes Chromosomes Cancer. 1998. PMID: 9559346 Review.
Cited by
- Biologic tumor behavior in pilocytic astrocytomas.
Belirgen M, Berrak SG, Ozdag H, Bozkurt SU, Eksioglu-Demiralp E, Ozek MM. Belirgen M, et al. Childs Nerv Syst. 2012 Mar;28(3):375-89. doi: 10.1007/s00381-011-1676-6. Epub 2012 Jan 14. Childs Nerv Syst. 2012. PMID: 22246337 - MAPK pathway activation in pilocytic astrocytoma.
Jones DT, Gronych J, Lichter P, Witt O, Pfister SM. Jones DT, et al. Cell Mol Life Sci. 2012 Jun;69(11):1799-811. doi: 10.1007/s00018-011-0898-9. Epub 2011 Dec 13. Cell Mol Life Sci. 2012. PMID: 22159586 Free PMC article. Review. - Genome-wide analysis of subependymomas shows underlying chromosomal copy number changes involving chromosomes 6, 7, 8 and 14 in a proportion of cases.
Kurian KM, Jones DT, Marsden F, Openshaw SW, Pearson DM, Ichimura K, Collins VP. Kurian KM, et al. Brain Pathol. 2008 Oct;18(4):469-73. doi: 10.1111/j.1750-3639.2008.00148.x. Epub 2008 Apr 7. Brain Pathol. 2008. PMID: 18397339 Free PMC article. - Pilocytic astrocytoma: pathology, molecular mechanisms and markers.
Collins VP, Jones DT, Giannini C. Collins VP, et al. Acta Neuropathol. 2015 Jun;129(6):775-88. doi: 10.1007/s00401-015-1410-7. Epub 2015 Mar 20. Acta Neuropathol. 2015. PMID: 25792358 Free PMC article. Review. - Mitogenic and progenitor gene programmes in single pilocytic astrocytoma cells.
Reitman ZJ, Paolella BR, Bergthold G, Pelton K, Becker S, Jones R, Sinai CE, Malkin H, Huang Y, Grimmet L, Herbert ZT, Sun Y, Weatherbee JL, Alberta JA, Daley JF, Rozenblatt-Rosen O, Condurat AL, Qian K, Khadka P, Segal RA, Haas-Kogan D, Filbin MG, Suva ML, Regev A, Stiles CD, Kieran MW, Goumnerova L, Ligon KL, Shalek AK, Bandopadhayay P, Beroukhim R. Reitman ZJ, et al. Nat Commun. 2019 Aug 19;10(1):3731. doi: 10.1038/s41467-019-11493-2. Nat Commun. 2019. PMID: 31427603 Free PMC article.
References
- Office for National Statistics The Health of Children and Young People. Mar 31, 2004. [Accessed June 2, 2005]. Available at: www.statistics.gov.uk/children/
- Kleihues P, Cavenee WK. Pathology and Genetics of Tumours of the Nervous System: World Health Organization Classification of Tumours. IARC Press; Lyon, France: 2000.
- Central Brain Tumour Registry of the United States . Statistical Report: Primary Brain Tumours in the United States, 1997–2001. CBTRUS; Chicago: 2005.
- Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. 2005;64:479–89. - PubMed
- Claus D, Sieber E, Engelhardt A, et al. Ascending central nervous spreading of a spinal astrocytoma. J Neurooncol. 1995;25:245–50. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous