Lethal giant discs, a novel C2-domain protein, restricts notch activation during endocytosis - PubMed (original) (raw)
Comparative Study
Lethal giant discs, a novel C2-domain protein, restricts notch activation during endocytosis
Jennifer L Childress et al. Curr Biol. 2006.
Abstract
The Notch signaling pathway plays a central role in animal growth and patterning, and its deregulation leads to many human diseases, including cancer. Mutations in the tumor suppressor lethal giant discs (lgd) induce strong Notch activation and hyperplastic overgrowth of Drosophila imaginal discs. However, the gene that encodes Lgd and its function in the Notch pathway have not yet been identified. Here, we report that Lgd is a novel, conserved C2-domain protein that regulates Notch receptor trafficking. Notch accumulates on early endosomes in lgd mutant cells and signals in a ligand-independent manner. This phenotype is similar to that seen when cells lose endosomal-pathway components such as Erupted and Vps25. Interestingly, Notch activation in lgd mutant cells requires the early endosomal component Hrs, indicating that Hrs is epistatic to Lgd. These data suggest that Lgd affects Notch trafficking between the actions of Hrs and the late endosomal component Vps25. Taken together, our data identify Lgd as a novel tumor-suppressor protein that regulates Notch signaling by targeting Notch for degradation or recycling.
Conflict of interest statement
Competing Interests statement The authors declare that they have no competing financial interests.
Figures
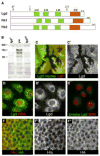
Figure 1. Lgd is a novel cytoplasmic C2 domain protein
(A) Schematic representation of the protein structures of Drosophila lgd and the two human homologs. The arrow labeled ‘d7’ indicates the position at which the lgdd7 allele has a frameshift mutation: the removal of a single nucleotide results in a new translational frame and a premature stop codon after the addition of 22 new amino acids. The arrow labeled ‘1’ indicates the position at which the lgd1 allele has a frameshift mutation: two nucleotides are removed resulting in a new translational frame and a premature stop codon after the addition of 15 new amino acids. The arrow labeled ‘J12’ indicates the position at which the EMS-induced allele, lgdJ12, contains a nonsense mutation: Q626 is changed to a stop codon. Green boxes indicate the Lgd homology domains (LH), and orange boxes indicate C2 domains. (B) Western blot to detect Lgd protein in extracts from lgd1, yw, and lgdd7 third instar larvae. The lgd1 and lgdd7 alleles truncate the protein N-terminally to the epitope used to generate the Lgd antibodies. The band at approximately 120kD contained the majority of Lgd protein, and was missing in both of the lgd mutant lanes. Other specific bands of lower molecular weight were also occasionally seen. (C–C′**)** Lgd antibodies specifically detect Lgd protein in imaginal discs. lgd mutant clones, indicated by the lack of GFP (green), stained with α–Lgd antibodies (red in C and gray in C′). (D–E) S2 cells stained with Lgd antibodies (green in D and E and gray in D′) and Topro (red). (E) Lgd antibodies incubated with Lgd antigen prior to use for staining. The preabsorbed Lgd antibodies (green) did not display specific staining, demonstrating that the Lgd antibodies specifically detect the Lgd antigen. (F–F″**)** C5 Gal4 driven UAS-HA-Lgd in the wing pouch of third instar imaginal discs, detected by α-HA (green in F and gray in F″). HA did not specifically colocalize with α-Hrs (red in F and gray in F′).
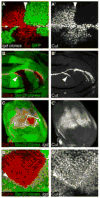
Figure 2. Lgd acts cells autonomously and restricts Notch activation in a ligand-independent manner
(A and A′**)** Third instar wing disc with lgdd10 mutant clones marked by the absence of GFP (Green). The discs were stained for Cut (red in A, gray in A′), a target of the Notch pathway. The arrow in (A) indicates the wing margin. The arrowhead in (A and A′) marks the clone boundary, showing the demarcation between Cut-expressing and non-expressing cells. Only lgd mutant cells (lacking GFP) showed Cut activation outside of the wing margin. (B–B′**)** Dlrev10,SerRX106 mutant clones in a wild-type wing disc. Cut (red in B and gray in B′) was ectopically expressed in some cells adjacent to the clone boundaries. (C–D′**)** Dlrev10,SerRX106 mutant clones in an lgd1/d7 wing imaginal disc. D and D′ show higher magnification of the region boxed in (C). The disc is stained for Cut (red in C and D, shown singly in gray in C′ and D′), and the lack of GFP expression (green in C and D) marks the clone. (D–D′) Cut expression extended throughout the clone. Also, the levels of Cut expression were higher along the boundary of the clone (arrowhead), demonstrating the presence of cis-inhibition in the absence of Lgd.
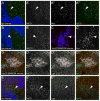
Figure 3. Mutations in lgd cause disruptions in Notch trafficking
(A–A‴**)** An lgdd7 mutant clone in a third instar wing disc, indicated by the absence of ubiGFP (blue), stained for α-NICD (green in A and A‴, gray in A′) and α-Hrs (red in A and A‴, gray in A″). NICD and Hrs accumulated together in lgd mutant cells (arrowhead). (B–B′**)** An lgdd7 mutant clone in a third instar wing disc, indicated by the absence of ubiGFP (blue) stained for α-NECD (green in B, gray in B′). The arrowhead indicates one of the subcellular puncta that shows accumulation of the extracellular part of the Notch protein. (C–C′**)** vps25, UAS-P35 MARCM mutant clones in the third instar wing disc, indicated by the presence of UAS-GFP (blue), stained for α-Hrs (red in C, gray in C′). The arrowhead indicates one of the subcellular puncta that expressed Hrs. (D–D‴**)** vps25 MARCM mutant clones in a third instar wing disc, indicated by the presence of UAS-GFP (blue) stained for α-NICD (green in D and D‴, gray in D′) and α-Lgd (red in D and D‴, gray in D″). Both NICD and Lgd protein accumulate in the vps25 mutant cells. (E–E′**)** An hrsD28 mutant clone in a third instar wing disc, indicated by the absence of ubiGFP (blue), stained for α-NICD (green in E and E‴ and gray in E′) and α-Lgd (red in E and E‴ gray in E″). NICD staining accumulated in the mutant cells, but there was no change in Lgd protein levels or localization within the clone compared to wild-type cells.
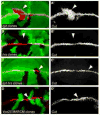
Figure 4. Hrs is required for Notch activation in lgd mutant cells
(A–A′**)** lgdd7 mutant clones in a third instar imaginal wing disc, indicated by the absence of ubiGFP (green), stained for Cut (red in A and gray in A′). Cut, a target gene activated by the Notch pathway, was induced in the lgd mutant tissue in the area surrounding the wing margin. The ectopic Cut induction does not appear to be autonomous in these clones only because this image and all panels in this figure are extended focus views of the whole disc, instead of single focal planes as in Fig. 1D. (B–B′**)** hrsD28 mutant clones in a third instar imaginal wing disc, indicated by the absence of ubiGFP (green), stained for α-Cut (red in B and gray in B′). Unlike in lgd mutant clones, Cut was not induced in the hrs mutant tissue. (C–C′**)** hrsD28, lgdd7 double mutant clones in a third instar imaginal wing disc, indicated by the absence of ubiGFP (green), stained for α-Cut (red in C and gray in C′). Cut was not induced in the cells lacking both Lgd and Hrs, suggesting that the hrs mutant phenotype is epistatic over the lgd mutant phenotype. (D–D′**)** vps25 MARCM mutant clones in the third instar wing disc, indicated by the presence of nuclear UAS-GFP (green), stained for α-Cut (red in D and gray in D′). Cut was not induced in the mutant tissue.
Similar articles
- The Drosophila Notch inhibitor and tumor suppressor gene lethal (2) giant discs encodes a conserved regulator of endosomal trafficking.
Jaekel R, Klein T. Jaekel R, et al. Dev Cell. 2006 Nov;11(5):655-69. doi: 10.1016/j.devcel.2006.09.019. Dev Cell. 2006. PMID: 17084358 - The conserved c2 domain protein lethal (2) giant discs regulates protein trafficking in Drosophila.
Gallagher CM, Knoblich JA. Gallagher CM, et al. Dev Cell. 2006 Nov;11(5):641-53. doi: 10.1016/j.devcel.2006.09.014. Dev Cell. 2006. PMID: 17084357 - Activation of Notch in lgd mutant cells requires the fusion of late endosomes with the lysosome.
Schneider M, Troost T, Grawe F, Martinez-Arias A, Klein T. Schneider M, et al. J Cell Sci. 2013 Jan 15;126(Pt 2):645-56. doi: 10.1242/jcs.116590. Epub 2012 Nov 23. J Cell Sci. 2013. PMID: 23178945 - Unravelling of Hidden Secrets: The Tumour Suppressor Lethal (2) Giant Discs (Lgd)/CC2D1, Notch Signalling and Cancer.
Reiff T, Baeumers M, Tibbe C, Klein T. Reiff T, et al. Adv Exp Med Biol. 2021;1287:31-46. doi: 10.1007/978-3-030-55031-8_3. Adv Exp Med Biol. 2021. PMID: 33034024 Review. - Endocytic Trafficking of the Notch Receptor.
Schnute B, Troost T, Klein T. Schnute B, et al. Adv Exp Med Biol. 2018;1066:99-122. doi: 10.1007/978-3-319-89512-3_6. Adv Exp Med Biol. 2018. PMID: 30030824 Review.
Cited by
- Balanced ubiquitination determines cellular responsiveness to extracellular stimuli.
Mukai A, Yamamoto-Hino M, Komada M, Okano H, Goto S. Mukai A, et al. Cell Mol Life Sci. 2012 Dec;69(23):4007-16. doi: 10.1007/s00018-012-1084-4. Epub 2012 Jul 24. Cell Mol Life Sci. 2012. PMID: 22825661 Free PMC article. Review. - Autocrine VEGF signaling is required for vascular homeostasis.
Lee S, Chen TT, Barber CL, Jordan MC, Murdock J, Desai S, Ferrara N, Nagy A, Roos KP, Iruela-Arispe ML. Lee S, et al. Cell. 2007 Aug 24;130(4):691-703. doi: 10.1016/j.cell.2007.06.054. Cell. 2007. PMID: 17719546 Free PMC article. - Endocytosis and intracellular trafficking of Notch and its ligands.
Yamamoto S, Charng WL, Bellen HJ. Yamamoto S, et al. Curr Top Dev Biol. 2010;92:165-200. doi: 10.1016/S0070-2153(10)92005-X. Curr Top Dev Biol. 2010. PMID: 20816395 Free PMC article. Review. - Structural Analysis of the ESCRT-III Regulator Lethal(2) Giant Discs/Coiled-Coil and C2 Domain-Containing Protein 1 (Lgd/CC2D1).
Breuer T, Tibbe C, Troost T, Klein T. Breuer T, et al. Cells. 2024 Jul 10;13(14):1174. doi: 10.3390/cells13141174. Cells. 2024. PMID: 39056756 Free PMC article. - Cc2d1b Contributes to the Regulation of Developmental Myelination in the Central Nervous System.
Acheta J, Hong J, Jeanette H, Brar S, Yalamanchili A, Feltri ML, Manzini MC, Belin S, Poitelon Y. Acheta J, et al. Front Mol Neurosci. 2022 May 3;15:881571. doi: 10.3389/fnmol.2022.881571. eCollection 2022. Front Mol Neurosci. 2022. PMID: 35592111 Free PMC article.
References
- Schweisguth F. Notch signaling activity. Curr Biol. 2004;14:R129–138. - PubMed
- Miele L. Notch signaling. Clin Cancer Res. 2006;12:1074–1079. - PubMed
- Agrawal N, Joshi S, Kango M, Saha D, Mishra A, Sinha P. Epithelial hyperplasia of imaginal discs induced by mutations in Drosophila tumor suppressor genes: growth and pattern formation in genetic mosaics. Dev Biol. 1995;169:387–398. - PubMed
- Klein T. The tumour suppressor gene l(2)giant discs is required to restrict the activity of Notch to the dorsoventral boundary during Drosophila wing development. Dev Biol. 2003;255:313–333. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM067997-03/GM/NIGMS NIH HHS/United States
- R01 GM067997-04/GM/NIGMS NIH HHS/United States
- T32 HD007325/HD/NICHD NIH HHS/United States
- T32-HD07325/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous