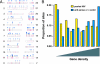Gene transfer in humans using a conditionally replicating lentiviral vector - PubMed (original) (raw)
. 2006 Nov 14;103(46):17372-7.
doi: 10.1073/pnas.0608138103. Epub 2006 Nov 7.
Laurent M Humeau, Jean Boyer, Rob-Roy MacGregor, Tessio Rebello, Xiaobin Lu, Gwendolyn K Binder, Vladimir Slepushkin, Franck Lemiale, John R Mascola, Frederic D Bushman, Boro Dropulic, Carl H June
Affiliations
- PMID: 17090675
- PMCID: PMC1635018
- DOI: 10.1073/pnas.0608138103
Gene transfer in humans using a conditionally replicating lentiviral vector
Bruce L Levine et al. Proc Natl Acad Sci U S A. 2006.
Abstract
We report findings from a clinical evaluation of lentiviral vectors in a phase I open-label nonrandomized clinical trial for HIV. This trial evaluated the safety of a conditionally replicating HIV-1-derived vector expressing an antisense gene against the HIV envelope. Five subjects with chronic HIV infection who had failed to respond to at least two antiviral regimens were enrolled. A single i.v. infusion of gene-modified autologous CD4 T cells was well tolerated in all patients. Viral loads were stable, and one subject exhibited a sustained decrease in viral load. CD4 counts remained steady or increased in four subjects, and sustained gene transfer was observed. Self-limiting mobilization of the vector was observed in four of five patients. There is no evidence for insertional mutagenesis after 21-36 months of observation. Immune function improved in four subjects. Lentiviral vectors appear promising for gene transfer to humans.
Conflict of interest statement
Conflict of interest statement: L.M.H., T.R., X.L., G.K.B., V.S., F.L., and B.D. were employees of VIRxSYS Corporation at the time this study was performed.
Figures
Fig. 1.
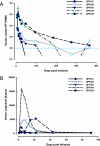
Schematic representation of the gene-transfer vector, VRX496, and viral loads and CD4 counts after treatment. (A) Vector design. (Upper) Schematic representation of pN1cptASenv (VRX496), depicting elements of the vector and the regions of wt-HIVNL4-3 from which they were derived. The numbers in the vector refer to the size of the genetic elements. VRX496 is derived from the NL4-3 clone of wild-type HIV. The vector expresses a 937-bp antisense segment targeted against HIV envelope gene (ASenv). The antisense payload is Tat- and Rev-dependent, and, thus, basal expression is increased after HIV infects vector-containing cells. HIV-derived elements include the 5′ and 3′ long terminal repeat (LTR), a packaging signal (Ψ), tRNA primer-binding site (pbs), central polypurine tract and central termination sequence (cPPT and CTS), splice acceptor and donor sites (SA and SD), Tat-dependent HIV promoter (P), Gag gene, rev response element (RRE), and 3′ polypurine tract (PPT). Engineered elements include a stop codon in gag (3). Gtag is a noncoding marker sequence from GFP. (Lower) Schema of pVRX577 (VIRPAC), the helper packaging construct. VRX496 is pseudotyped with a vesicular stomatitis virus protein G (VSV-G) envelope. Gag and pol are expressed under the control of the CMV promoter, Rev under the control of the rev response element derived from HIV-2 (RRE-2), which is used to reduce homology between VRX496 and VIRPAC, tat under the control of an internal ribosomal entry site (IRES), and VSV-G expressed by an elongation factor 1α (EF-1α)/human T cell lymphotrophic virus (HTLV) chimeric promoter. VSV-G is separated from the other packaging genes for safety by several pause signals and a _cis_-acting ribozyme derived from the tobacco mosaic ringspot virus (sTobRV+Rz) that will cleave any read-through RNA. Sequences of rev and tat genes were partially degenerated to reduce homology with the vector. (B) Primary endpoints. The log plasma HIV viral load is depicted as change from baseline. The baseline values are shown in Table 1. Changes >0.5 log were outside the variation of the assay and were considered meaningful. The VRX496 cell infusion was given on day 0. Subject 4 began an antiretroviral therapy regimen 7 months after infusion, and his viral load became undetectable at that point. (C) Subject 2 course and detailed viral load. The detailed course for subject 2 is shown, with all available viral loads and a summary of antiretroviral therapy plotted. Note that the _x_-axis scale changes on day 0 to display the available baseline values. The vertical arrow depicts the time of the VRX 496 modified CD4+ cell infusion. The date of infusion was October 13, 2003; the most recent viral load is 1,930 copies per ml (January 2006). (D) CD4 cell counts. CD4 cell counts are plotted as a change from baseline after the VRX496 infusion. Baseline values are shown in Table 1. Subject 4 began a new antiretroviral therapy regimen 7 months after infusion, and his viral load became undetectable at that point.
Fig. 2.
Engraftment of vector-modified cells and detection of mobilization of vector in vivo. (A) Prolonged engraftment of lentiviral transduced CD4+ cells. Vector persistence was assessed beginning 20 min after infusion of VRX496-modified CD4+ cells and then at 72 h; 1, 2, 3, and 6 weeks; and 3, 6, 9, and 12 months. PBMC were collected at the indicated time points, and DNA analysis was performed for detection of VRX496 vector sequences by using real-time PCR. The limit of quantitative detection (LOD) is 200 vector copies per 106 PBMC. At the 1-year time point, subject 4 has a frequency of engraftment of 0.04% (400 copies) after being undetectable at the 9-month time point, and subject 2 has 0.023% engraftment (233 copies) at 2 years. Subjects 3–5 have not yet completed their 2-year follow up. (B) Vector mobilization. To assess for vector mobilization, RT-PCR analysis for vector genomic RNA was done by using primers specific for truncated GFP (Gtag) sequence, which is a component of the vector and allowed it to be distinguished from wt-HIV in the patients (see Methods). The presence of genomic vector transcripts in circulation was assessed by isolating RNA from plasma at the indicated time points after infusion on day 0.
Fig. 3.
Analysis of the sites of VRX496 integration in patient cell product prior to infusion. (A) Integration of the VRX496 antisense env vector in the human genome. Integration sites are mapped on the human chromosomes, with sites of vector integration shown by the blue “lollipops.” Gene density is indicated by the red shading on the human chromosomes, with more intense red indicating higher gene density. Vector integration differs from random (P = 2 × 10−10 for comparison with random placement). (B) Preferential integration of the VRX496 antisense env vector in gene-rich regions. The association of integration sites with genes was assessed by sliding a 250-kb window along the human genome. Values were determined separately for antisense env vector sites and summed previously studied HIV sites. The results were pooled, divided into nine intervals, and the proportion of each type of site in each interval assessed. Vector integration in gene-dense regions was more strongly favored than in previously studied HIV vector or virus data sets (P = 2.9 ×10−5 for comparison with pooled HIV integration data). The P value is the result of fitting a cubic polynomial to the gene-density values. See the online statistical supplement (Appendix) for more details.
Fig. 4.
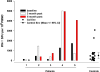
Immunologic assessment of HIV-1 _env_-specific effector cells secreting IFN- γ. Blood samples were obtained at baseline and at 3 and 6 months after gene-transfer therapy. PBMCs were isolated from study subjects and HIV-1-positive control subjects (n = 25) by a standard Ficoll separation technique. IFN-γ production after HIV-1 env in vitro stimulation of PBMCs was assessed for an env antigen-specific response by a standard ELISPOT. The mean ± 95% confidence interval for the control subjects is plotted. ∗, subject 1 did not have a 6-month sample available for analysis.
Similar articles
- Lentiviral vectors ready for prime-time.
Kohn DB. Kohn DB. Nat Biotechnol. 2007 Jan;25(1):65-6. doi: 10.1038/nbt0107-65. Nat Biotechnol. 2007. PMID: 17211402 No abstract available. - Antiviral effects of autologous CD4 T cells genetically modified with a conditionally replicating lentiviral vector expressing long antisense to HIV.
Tebas P, Stein D, Binder-Scholl G, Mukherjee R, Brady T, Rebello T, Humeau L, Kalos M, Papasavvas E, Montaner LJ, Schullery D, Shaheen F, Brennan AL, Zheng Z, Cotte J, Slepushkin V, Veloso E, Mackley A, Hwang WT, Aberra F, Zhan J, Boyer J, Collman RG, Bushman FD, Levine BL, June CH. Tebas P, et al. Blood. 2013 Feb 28;121(9):1524-33. doi: 10.1182/blood-2012-07-447250. Epub 2012 Dec 20. Blood. 2013. PMID: 23264589 Free PMC article. Clinical Trial. - Efficient lentiviral vector-mediated control of HIV-1 replication in CD4 lymphocytes from diverse HIV+ infected patients grouped according to CD4 count and viral load.
Humeau LM, Binder GK, Lu X, Slepushkin V, Merling R, Echeagaray P, Pereira M, Slepushkina T, Barnett S, Dropulic LK, Carroll R, Levine BL, June CH, Dropulic B. Humeau LM, et al. Mol Ther. 2004 Jun;9(6):902-13. doi: 10.1016/j.ymthe.2004.03.005. Mol Ther. 2004. PMID: 15194057 - Gene therapy of HIV-1 infection using lentiviral vectors expressing anti-HIV-1 genes.
Mautino MR, Morgan RA. Mautino MR, et al. AIDS Patient Care STDS. 2002 Jan;16(1):11-26. doi: 10.1089/108729102753429361. AIDS Patient Care STDS. 2002. PMID: 11839215 Review. - Production and titration of lentiviral vectors.
Salmon P, Trono D. Salmon P, et al. Curr Protoc Neurosci. 2006 Nov;Chapter 4:Unit 4.21. doi: 10.1002/0471142301.ns0421s37. Curr Protoc Neurosci. 2006. PMID: 18428637 Review.
Cited by
- RD-MolPack technology for the constitutive production of self-inactivating lentiviral vectors pseudotyped with the nontoxic RD114-TR envelope.
Marin V, Stornaiuolo A, Piovan C, Corna S, Bossi S, Pema M, Giuliani E, Scavullo C, Zucchelli E, Bordignon C, Rizzardi GP, Bovolenta C. Marin V, et al. Mol Ther Methods Clin Dev. 2016 May 11;3:16033. doi: 10.1038/mtm.2016.33. eCollection 2016. Mol Ther Methods Clin Dev. 2016. PMID: 27222840 Free PMC article. - A TLR and non-TLR mediated innate response to lentiviruses restricts hepatocyte entry and can be ameliorated by pharmacological blockade.
Agudo J, Ruzo A, Kitur K, Sachidanandam R, Blander JM, Brown BD. Agudo J, et al. Mol Ther. 2012 Dec;20(12):2257-67. doi: 10.1038/mt.2012.150. Epub 2012 Aug 7. Mol Ther. 2012. PMID: 22871668 Free PMC article. - Characterization of a third generation lentiviral vector pseudotyped with Nipah virus envelope proteins for endothelial cell transduction.
Witting SR, Vallanda P, Gamble AL. Witting SR, et al. Gene Ther. 2013 Oct;20(10):997-1005. doi: 10.1038/gt.2013.23. Epub 2013 May 23. Gene Ther. 2013. PMID: 23698741 Free PMC article. - Adoptive cell therapy: genetic modification to redirect effector cell specificity.
Morgan RA, Dudley ME, Rosenberg SA. Morgan RA, et al. Cancer J. 2010 Jul-Aug;16(4):336-41. doi: 10.1097/PPO.0b013e3181eb3879. Cancer J. 2010. PMID: 20693844 Free PMC article. Review. - Recent advances in lentiviral vector development and applications.
Mátrai J, Chuah MK, VandenDriessche T. Mátrai J, et al. Mol Ther. 2010 Mar;18(3):477-90. doi: 10.1038/mt.2009.319. Epub 2010 Jan 19. Mol Ther. 2010. PMID: 20087315 Free PMC article. Review.
References
- Levine BL, Mosca J, Riley JL, Carroll RG, Vahey MT, Jagodizinski L, Wagner KF, Mayers DL, Burke DS, Weislow OS, et al. Science. 1996;272:1939–1943. - PubMed
- Lu X, Humeau L, Slepushkin V, Binder G, Yu Q, Slepushkina T, Chen Z, Merling R, Davis B, Chang YN, et al. J Gene Med. 2004;6:963–973. - PubMed
- Levine BL, Bernstein WB, Aronson NE, Schlienger K, Cotte J, Perfetto S, Humphries MJ, Ratto-Kim S, Birx DL, Steffans C, et al. Nat Med. 2002;8:47–53. - PubMed
- Humeau LM, Binder GK, Lu X, Slepushkin V, Merling R, Echeagaray P, Pereira M, Slepushkina T, Barnett S, Dropulic LK, et al. Mol Ther. 2004;9:902–913. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2R44AI051908/AI/NIAID NIH HHS/United States
- U19-AI066290/AI/NIAID NIH HHS/United States
- U19 AI066290/AI/NIAID NIH HHS/United States
- R44 AI051908/AI/NIAID NIH HHS/United States
- U19 AI066290-010001/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials