The frequency of translational misreading errors in E. coli is largely determined by tRNA competition - PubMed (original) (raw)
The frequency of translational misreading errors in E. coli is largely determined by tRNA competition
Emily B Kramer et al. RNA. 2007 Jan.
Abstract
Estimates of missense error rates (misreading) during protein synthesis vary from 10(-3) to 10(-4) per codon. The experiments reporting these rates have measured several distinct errors using several methods and reporter systems. Variation in reported rates may reflect real differences in rates among the errors tested or in sensitivity of the reporter systems. To develop a more accurate understanding of the range of error rates, we developed a system to quantify the frequency of every possible misreading error at a defined codon in Escherichia coli. This system uses an essential lysine in the active site of firefly luciferase. Mutations in Lys529 result in up to a 1600-fold reduction in activity, but the phenotype varies with amino acid. We hypothesized that residual activity of some of the mutant genes might result from misreading of the mutant codons by tRNA(Lys) (UUUU), the cognate tRNA for the lysine codons, AAA and AAG. Our data validate this hypothesis and reveal details about relative missense error rates of near-cognate codons. The error rates in E. coli do, in fact, vary widely. One source of variation is the effect of competition by cognate tRNAs for the mutant codons; higher error frequencies result from lower competition from low-abundance tRNAs. We also used the system to study the effect of ribosomal protein mutations known to affect error rates and the effect of error-inducing antibiotics, finding that they affect misreading on only a subset of near-cognate codons and that their effect may be less general than previously thought.
Figures
FIGURE 1.
Near- and noncognate mutations for Lysine-529. The two Lys codons are shown in italics in this standard genetic code table. Near-cognate codons, differing at one codon position, are shown in reverse on black. Noncognate synonymous codons, differing in more than one codon position, are shown in black on gray. The control UUU codon is shown in boldface.
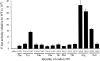
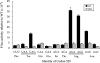
FIGURE 2.
Variation in F-luc activity of K5290 mutants. The graph shows the F-luc expression of the indicated constructs expressed as a fraction of the expression of wild-type F-luc. The mutants include a deletion of the entire firefly luc gene (Fluc) and those replacing the K529 codon (AAA) with the indicated codon. Indicated below each codon is the amino acid it encodes. Error bars are standard errors of the mean.
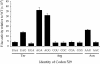
FIGURE 3.
High residual F-luc activity of some K529 codons results from near-cognate decoding. A comparison of the F-luc activity relative to wild type of synonymous near- and noncognate codon mutants. Codon identity is shown as in Figure 2.
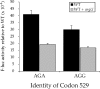
FIGURE 4.
Overexpression of tRNAArg UCU reduces misreading errors at Arg codons. The F-luc activity relative to wild type is shown for mutants replacing the K529 codon with the Arg codons AGA or AGG. Black bars indicate expression in a wild-type genetic background (WT) and gray bars indicate expression in the presence of overexpressed tRNAArg UCU.
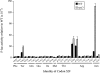
FIGURE 5.
Aminoglycoside antibiotics increase misreading on a subset of near-cognate codons. The F-luc activities relative to wild type are shown for mutants carrying the indicated K529 codon replacements in the presence of no antibiotic (black bars), 5 μg/mL paromomycin (gray bars), or 2 μg/mL streptomycin (white bars).
FIGURE 6.
A mutation in rpsD increases misreading only of error-prone codons. The F-luc activities relative to wild type of the indicated K529 codon replacements are shown from a wild-type strain (black bars) and a strain carrying the rpsD12 mutation (gray bars).
FIGURE 7.
A mutation in rpsL reduces misreading only of error-prone codons. The F-luc activities relative to wild type of the indicated K529 codon replacements are shown from a wild-type strain (black bars) and a strain carrying the rpsL141 mutation (gray bars).
Similar articles
- A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae.
Kramer EB, Vallabhaneni H, Mayer LM, Farabaugh PJ. Kramer EB, et al. RNA. 2010 Sep;16(9):1797-808. doi: 10.1261/rna.2201210. Epub 2010 Jul 22. RNA. 2010. PMID: 20651030 Free PMC article. - Translational misreading in Mycobacterium smegmatis increases in stationary phase.
Leng T, Pan M, Xu X, Javid B. Leng T, et al. Tuberculosis (Edinb). 2015 Dec;95(6):678-681. doi: 10.1016/j.tube.2015.09.010. Epub 2015 Oct 19. Tuberculosis (Edinb). 2015. PMID: 26542220 - Differentiating between near- and non-cognate codons in Saccharomyces cerevisiae.
Plant EP, Nguyen P, Russ JR, Pittman YR, Nguyen T, Quesinberry JT, Kinzy TG, Dinman JD. Plant EP, et al. PLoS One. 2007 Jun 13;2(6):e517. doi: 10.1371/journal.pone.0000517. PLoS One. 2007. PMID: 17565370 Free PMC article. - The problem of genetic code misreading during protein synthesis.
Joshi K, Cao L, Farabaugh PJ. Joshi K, et al. Yeast. 2019 Jan;36(1):35-42. doi: 10.1002/yea.3374. Yeast. 2019. PMID: 30557461 Review. - The ribosome's response to codon-anticodon mismatches.
Daviter T, Gromadski KB, Rodnina MV. Daviter T, et al. Biochimie. 2006 Aug;88(8):1001-11. doi: 10.1016/j.biochi.2006.04.013. Epub 2006 May 12. Biochimie. 2006. PMID: 16716484 Review.
Cited by
- Association analysis of DACT1 genetic variants and gastric cancer risk in a Chinese Han population: a case-control study.
Huang C, Wang Y, Fan H, Ma X, Tang R, Huan X, Zhu Y, Xu Z, Xu H, Yang L. Huang C, et al. Onco Targets Ther. 2016 Sep 29;9:5975-5983. doi: 10.2147/OTT.S109899. eCollection 2016. Onco Targets Ther. 2016. PMID: 27729806 Free PMC article. - Genetic code translation displays a linear trade-off between efficiency and accuracy of tRNA selection.
Johansson M, Zhang J, Ehrenberg M. Johansson M, et al. Proc Natl Acad Sci U S A. 2012 Jan 3;109(1):131-6. doi: 10.1073/pnas.1116480109. Epub 2011 Dec 21. Proc Natl Acad Sci U S A. 2012. PMID: 22190491 Free PMC article. - Transfer RNAs: diversity in form and function.
Berg MD, Brandl CJ. Berg MD, et al. RNA Biol. 2021 Mar;18(3):316-339. doi: 10.1080/15476286.2020.1809197. Epub 2020 Sep 9. RNA Biol. 2021. PMID: 32900285 Free PMC article. Review. - Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome.
Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Neumann H, et al. Nature. 2010 Mar 18;464(7287):441-4. doi: 10.1038/nature08817. Epub 2010 Feb 14. Nature. 2010. PMID: 20154731 - Modulating Mistranslation Potential of tRNASer in Saccharomyces cerevisiae.
Berg MD, Zhu Y, Genereaux J, Ruiz BY, Rodriguez-Mias RA, Allan T, Bahcheli A, Villén J, Brandl CJ. Berg MD, et al. Genetics. 2019 Nov;213(3):849-863. doi: 10.1534/genetics.119.302525. Epub 2019 Sep 4. Genetics. 2019. PMID: 31484688 Free PMC article.
References
- Amann, E., Ochs, B., Abel, K.J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli . Gene. 1988;69:301–315. - PubMed
- Andersson, D.I., Bohman, K., Isaksson, L.A., Kurland, C.G. Translation rates and misreading characteristics of rpsD mutants in Escherichia coli . Mol. Gen. Genet. 1982;187:467–472. - PubMed
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., Struhl, K. Current protocols in molecular biology. John Wiley & Sons; New York: 1997.
- Branchini, B.R., Murtiashaw, M.H., Magyar, R.A., Anderson, S.M. The role of lysine 529, a conserved residue of the acyl-adenylate-forming enzyme superfamily, in firefly luciferase. Biochemistry. 2000;39:5433–5440. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources