Microarray-based analysis of microbial community RNAs by whole-community RNA amplification - PubMed (original) (raw)
Microarray-based analysis of microbial community RNAs by whole-community RNA amplification
Haichun Gao et al. Appl Environ Microbiol. 2007 Jan.
Abstract
A new approach, termed whole-community RNA amplification (WCRA), was developed to provide sufficient amounts of mRNAs from environmental samples for microarray analysis. This method employs fusion primers (six to nine random nucleotides with an attached T7 promoter) for the first-strand synthesis. The shortest primer (T7N6S) gave the best results in terms of the yield and representativeness of amplification. About 1,200- to 1,800-fold amplification was obtained with amounts of the RNA templates ranging from 10 to 100 ng, and very representative detection was obtained with 50 to 100 ng total RNA. Evaluation with a Shewanella oneidensis Deltafur strain revealed that the amplification method which we developed could preserve the original abundance relationships of mRNAs. In addition, to determine whether representative detection of RNAs can be achieved with mixed community samples, amplification biases were evaluated with a mixture containing equal quantities of RNAs (100 ng each) from four bacterial species, and representative amplification was also obtained. Finally, the method which we developed was applied to the active microbial populations in a denitrifying fluidized bed reactor used for denitrification of contaminated groundwater and ethanol-stimulated groundwater samples for uranium reduction. The genes expressed were consistent with the expected functions of the bioreactor and groundwater system, suggesting that this approach is useful for analyzing the functional activities of microbial communities. This is one of the first demonstrations that microarray-based technology can be used to successfully detect the activities of microbial communities from real environmental samples in a high-throughput fashion.
Figures
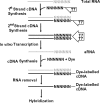
FIG. 1.
Outline of the WCRA method. Gray type, RNA; black type, DNA. The primers used and other details are described in the text.
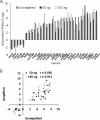
FIG. 2.
Evaluation of amplification biases using whole-genome S. oneidensis cDNA microarrays. (A) Primers tested in this study. N indicates a random nucleotide. (B) Ratios (amplified/unamplified) were plotted against the order of the genes in the genome, from SO0001 to SOA0173. In the unamplified panel, hybridization was carried out with the same unamplified RNA labeled with Cy3 and Cy5. In all other panels, 500 ng of RNA was amplified using WCRA with the primers indicated, labeled with Cy5, and hybridized with unamplified RNA (Cy3).
FIG. 3.
Scatter plots of genes from replicate hybridizations and replicate amplification reactions. (A to C) Scatter plots comparing the expression profiles of unamplified RNA from the wild-type strain (wt) with the expression profiles of unamplified RNA (A), 100 ng amplified starting RNA (B), or 50 ng amplified starting RNA (C) from the Δ_fur_ strain. (D) Quantitative analysis of relationships for expression ratios of all genes (Δ_fur_ RNA/wild-type RNA) between the amplified RNA (100 ng) and the unamplified RNA.
FIG. 4.
Amplification bias analysis by expression ratio comparison. All 30 genes examined have been reported to be highly affected by the fur mutation (36). (A) Expression ratios (Δ_fur_ RNA/wild-type RNA) for the genes obtained with unamplified RNA and 50 and 100 ng of the starting RNA. (B) Quantitative analysis of relationships for expression ratios of the 30 genes (Δ_fur_ RNA/wild-type RNA) between the amplified RNA and the unamplified RNA.
Similar articles
- Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification.
Wu L, Liu X, Schadt CW, Zhou J. Wu L, et al. Appl Environ Microbiol. 2006 Jul;72(7):4931-41. doi: 10.1128/AEM.02738-05. Appl Environ Microbiol. 2006. PMID: 16820490 Free PMC article. - Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis.
Kurimoto K, Yabuta Y, Ohinata Y, Saitou M. Kurimoto K, et al. Nat Protoc. 2007;2(3):739-52. doi: 10.1038/nprot.2007.79. Nat Protoc. 2007. PMID: 17406636 - Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation.
Brodie EL, Desantis TZ, Joyner DC, Baek SM, Larsen JT, Andersen GL, Hazen TC, Richardson PM, Herman DJ, Tokunaga TK, Wan JM, Firestone MK. Brodie EL, et al. Appl Environ Microbiol. 2006 Sep;72(9):6288-98. doi: 10.1128/AEM.00246-06. Appl Environ Microbiol. 2006. PMID: 16957256 Free PMC article. - DNA microarrays--techniques and applications in microbial systems.
Majtán T, Bukovská G, Timko J. Majtán T, et al. Folia Microbiol (Praha). 2004;49(6):635-64. doi: 10.1007/BF02931546. Folia Microbiol (Praha). 2004. PMID: 15881400 Review. - Options available for profiling small samples: a review of sample amplification technology when combined with microarray profiling.
Nygaard V, Hovig E. Nygaard V, et al. Nucleic Acids Res. 2006 Feb 9;34(3):996-1014. doi: 10.1093/nar/gkj499. Print 2006. Nucleic Acids Res. 2006. PMID: 16473852 Free PMC article. Review.
Cited by
- Microbial Diagnostic Array Workstation (MDAW): a web server for diagnostic array data storage, sharing and analysis.
Scaria J, Sreedharan A, Chang YF. Scaria J, et al. Source Code Biol Med. 2008 Sep 23;3:14. doi: 10.1186/1751-0473-3-14. Source Code Biol Med. 2008. PMID: 18811969 Free PMC article. - Warming Alters Expressions of Microbial Functional Genes Important to Ecosystem Functioning.
Xue K, Xie J, Zhou A, Liu F, Li D, Wu L, Deng Y, He Z, Van Nostrand JD, Luo Y, Zhou J. Xue K, et al. Front Microbiol. 2016 May 6;7:668. doi: 10.3389/fmicb.2016.00668. eCollection 2016. Front Microbiol. 2016. PMID: 27199978 Free PMC article. - Amplification of viral RNA from drinking water using TransPlex™ whole-transcriptome amplification.
Parker JK, Chang TY, Meschke JS. Parker JK, et al. J Appl Microbiol. 2011 Jul;111(1):216-23. doi: 10.1111/j.1365-2672.2011.05029.x. Epub 2011 May 5. J Appl Microbiol. 2011. PMID: 21477067 Free PMC article. - Functional Gene Array-Based Ultrasensitive and Quantitative Detection of Microbial Populations in Complex Communities.
Shi Z, Yin H, Van Nostrand JD, Voordeckers JW, Tu Q, Deng Y, Yuan M, Zhou A, Zhang P, Xiao N, Ning D, He Z, Wu L, Zhou J. Shi Z, et al. mSystems. 2019 Jun 18;4(4):e00296-19. doi: 10.1128/mSystems.00296-19. mSystems. 2019. PMID: 31213523 Free PMC article. - Using DNA microarrays to assay part function.
Rhodius VA, Gross CA. Rhodius VA, et al. Methods Enzymol. 2011;497:75-113. doi: 10.1016/B978-0-12-385075-1.00004-4. Methods Enzymol. 2011. PMID: 21601083 Free PMC article.
References
- Adamczyk, J., M. Hesselsoe, N. Iversen, M. Horn, A. Lehner, P. H. Nielsen, M. Schloter, P. Roslev, and M. Wagner. 2003. The isotope array, a new tool that employs substrate-mediated labeling of rRNA for determination of microbial community structure and function. Appl. Environ. Microbiol. 69:6875-6887. - PMC - PubMed
- Bodrossy, L., N. Stralis-Pavese, J. C. Murrell, S. Radajewski, A. Weilharter, and A. Sessitsch. 2003. Development and validation of a diagnostic microbial microarray for methanotrophs. Environ. Microbiol. 5:566-582. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources