Two-step recruitment of RNA-directed DNA methylation to tandem repeats - PubMed (original) (raw)
Two-step recruitment of RNA-directed DNA methylation to tandem repeats
Simon W-L Chan et al. PLoS Biol. 2006 Nov.
Abstract
Tandem repeat sequences are frequently associated with gene silencing phenomena. The Arabidopsis thaliana FWA gene contains two tandem repeats and is an efficient target for RNA-directed de novo DNA methylation when it is transformed into plants. We showed that the FWA tandem repeats are necessary and sufficient for de novo DNA methylation and that repeated character rather than intrinsic sequence is likely important. Endogenous FWA can adopt either of two stable epigenetic states: methylated and silenced or unmethylated and active. Surprisingly, we found small interfering RNAs (siRNAs) associated with FWA in both states. Despite this, only the methylated form of endogenous FWA could recruit further RNA-directed DNA methylation or cause efficient de novo methylation of transgenic FWA. This suggests that RNA-directed DNA methylation occurs in two steps: first, the initial recruitment of the siRNA-producing machinery, and second, siRNA-directed DNA methylation either in cis or in trans. The efficiency of this second step varies depending on the nature of the siRNA-producing locus, and at some loci, it may require pre-existing chromatin modifications such as DNA methylation itself. Enhancement of RNA-directed DNA methylation by pre-existing DNA methylation could create a self-reinforcing system to enhance the stability of silencing. Tandem repeats throughout the Arabidopsis genome produce siRNAs, suggesting that repeat acquisition may be a general mechanism for the evolution of gene silencing.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
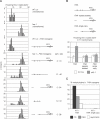
Figure 1. De Novo DNA Methylation of Transformed FWA Can Be Rescued by Wild-Type Genes from Pollen
Heterozygous rdr2–1, ago4–1, and drm1–1 drm2–1 plants were transformed with FWA. If the gametophytic genotype controls de novo DNA methylation and silencing, a 1:1 ratio of early- to late-flowering plants is predicted. If a wild-type gene from pollen can rescue de novo DNA methylation in the fertilized zygote, a 3:1 early- to late-flowering ratio results. The ratio of early- to late-flowering plants was determined in the T1 generation (see Figure S1 for determination of early versus late flowering in each ecotype). The probability of the observed results occurring by chance assuming a 1:1 or 3:1 ratio was calculated with the χ2 test.
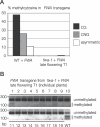
Figure 2. Recruitment of RNA-Directed DNA Methylation to Transformed Direct Repeats
(A) Silencing of endogenous FWA by transformed FWA requires tandem repeats in the incoming transgene. Wild-type (WT) L_er_ or fwa-1 plants were transformed with a complete FWA transgene or the deletion variants shown at right. FWA contains two tandem repeats, which are depicted as two sets of arrows. Parental fwa-1 plants are uniformly late flowering. Early flowering in fwa-1 transformants occurs when endogenous fwa is silenced by de novo DNA methylation. Plants were scored as early- (17 leaves or fewer) or late-flowering (18 leaves or more) by comparison to untransformed L_er_ and fwa-1. (B) FWA transgenes lacking tandem repeats do not cause late flowering when transformed into rdr2–1. The full FWA gene, “FWA repeats deleted” transgene, and “FWA single copy” transgenes are diagrammed. Wild-type Col and rdr2–1 plants were transformed with each transgene, and flowering time was assayed in the T1 generation. (C) De novo DNA methylation of transformed FWA requires tandem repeat character. Wild-type Col plants were transformed with full-length FWA or with single-copy FWA. DNA methylation of the FWA transgene was assayed by bisulfite sequencing.
Figure 3. Communication between Methylated Endogenous FWA and Transformed FWA
(A) Efficient de novo DNA methylation of transgenic FWA requires DNA methylation at endogenous FWA. DNA methylation of an FWA transgene introduced into wild-type or fwa-1 plants was assayed by bisulfite sequencing. For fwa-1 + FWA, three clones were sequenced from each of eight late-flowering individuals (each transformant had >20 rosette leaves at bolting). Early-flowering fwa-1 transformants were discarded, because they have silenced the FWA transgene by de novo DNA methylation. (B) Transgenic FWA is unmethylated in many late-flowering fwa-1 + FWA T1 individuals. Before bisulfite treatment, genomic DNA from late-flowering fwa + FWA T1 plants was digested with BglII to destroy the endogenous FWA gene. DNA methylation of transgenic FWA was assayed by PCR from bisulfite-treated DNA followed by ClaI digestion. CG DNA methylation protects the ClaI site from bisulfite conversion, allowing restriction digestion after bisulfite treatment. Wild-type DNA (not digested with BglII) was assayed as a control.
Figure 4. Characterization of FWA siRNAs in Different Genetic Backgrounds
(A) Schematic diagram of the FWA locus and position of LNA probe used to detect siRNAs. (B) FWA siRNAs are still produced when the gene is unmethylated. siRNAs from wild type, fwa-1, and met1–3 were analyzed by Northern blotting. miR159 was probed as a loading control. (C) FWA siRNAs from plants with the indicated genotype were analyzed by Northern blotting. (D) siRNAs in fwa-1 do not cause mRNA destruction. The ratio of FWA to ACTIN7 mRNA levels in rosette leaf tissue was measured by RT-qPCR. Reverse transcription was performed with a poly-T oligonucleotide. There was no PCR amplification when reverse transcriptase was omitted (unpublished data).
Figure 5. Separable Recruitment of siRNA Production and Non-CG DNA Methylation at Chromosomal Tandem Repeats
(A) Unmethylated chromosomal tandem repeats can recruit siRNA production. fwa-1 nrpd1a-1 and fwa-1 rdr2–1 lack FWA siRNAs, but when these mutants are crossed together, Northern blotting shows that FWA siRNA production is restored in F1 plants. (B) Unmethylated chromosomal tandem repeats cannot recruit de novo DNA methylation. When fwa-1 nrpd1a-1 and fwa-1 rdr2–1 mutants are crossed together, FWA remains unmethylated despite resumption of siRNA production. DNA methylation of FWA was assayed by PCR from bisulfite-treated DNA followed by ClaI digestion. (C) CG DNA methylation recruits siRNA-directed non-CG DNA methylation to FWA. When the recessive rdr2–1 and nrpd1a-1 mutants are crossed together, non-CG DNA methylation returns to CG-methylated FWA in the F1 plants. DNA methylation was assayed by bisulfite sequencing. (D) CG DNA methylation recruits siRNA-directed non-CG DNA methylation to MEA-ISR. Non-CG DNA methylation returns to CG-methylated MEA-ISR in rdr2–1 × nrpd1a-1 F1 plants. Asymmetric DNA methylation was assayed by PCR from bisulfite-treated DNA followed by BamHI digestion. DNA methylation protects the BamHI site from bisulfite conversion, allowing restriction digestion after bisulfite treatment.
Figure 6. Association of siRNAs with Tandem Repeats throughout the Arabidopsis Genome
(A) Chromosomal distribution of unique tandem repeats that produce siRNAs. Single-copy tandem repeats were identified (see text for details) and compared against a large database of cloned siRNAs [18]. The five Arabidopsis chromosomes are shown as rectangular boxes. Tandem repeats that overlap with dense or moderate siRNA clusters are shown as black dots (dots above the box represent euchromatic tandem repeats that were relatively close to each other). Pericentromeric heterochromatin is shown as gray shading. (B) Unique tandem repeats in euchromatin are enriched for siRNA production relative to randomly chosen sequences. Unique tandem repeats and a set of random sequences of similar size (141 bp is the median size of unique tandem repeats) were assessed for overlap with dense or moderate siRNA clusters. The probability of selecting 30/1494 siRNA-producing loci by chance (assuming the same siRNA-producing frequency as randomly chosen windows) was calculated with a one-sided binomial test. (C) DNA methylation of unique tandem repeats in euchromatin. Unique tandem repeats with and without siRNAs were compared to genome-wide DNA methylation data from an immunoprecipitation/tiling microarray experiment [37]. The fraction 221/1464 unique tandem repeats without siRNAs were methylated, compared with 14/30 unique tandem repeats that had siRNAs. The statistical significance of overlap between siRNA production and DNA methylation was calculated with a right-tailed hypergeometric test.
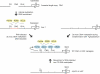
Figure 7. Model for RNA-Directed DNA Methylation at Tandem Repeats
An ancestral single-copy gene may undergo internal duplication, creating tandem repeats that recruit siRNA-producing factors. After initial siRNA production, downstream RNA-directed DNA methylation may or may not occur efficiently at a particular locus, resulting in stable methylated or unmethylated genes, both of which produce siRNAs (small arrows). Transformed FWA efficiently recruits both siRNA production and downstream de novo DNA methylation. CG DNA methylation allows recruitment of siRNA-directed DNA methylation, providing a self-reinforcing feedback loop at silent loci.
Comment in
- siRNAs and DNA methylation do a two-step to silence tandem sequences.
Robinson R. Robinson R. PLoS Biol. 2006 Nov;4(11):e407. doi: 10.1371/journal.pbio.0040407. Epub 2006 Oct 24. PLoS Biol. 2006. PMID: 20076502 Free PMC article. No abstract available.
Similar articles
- Distinct and concurrent pathways of Pol II- and Pol IV-dependent siRNA biogenesis at a repetitive trans-silencer locus in Arabidopsis thaliana.
Sasaki T, Lee TF, Liao WW, Naumann U, Liao JL, Eun C, Huang YY, Fu JL, Chen PY, Meyers BC, Matzke AJ, Matzke M. Sasaki T, et al. Plant J. 2014 Jul;79(1):127-38. doi: 10.1111/tpj.12545. Epub 2014 Jun 13. Plant J. 2014. PMID: 24798377 - Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats.
Zilberman D, Cao X, Johansen LK, Xie Z, Carrington JC, Jacobsen SE. Zilberman D, et al. Curr Biol. 2004 Jul 13;14(13):1214-20. doi: 10.1016/j.cub.2004.06.055. Curr Biol. 2004. PMID: 15242620 - Specific tandem repeats are sufficient for paramutation-induced trans-generational silencing.
Belele CL, Sidorenko L, Stam M, Bader R, Arteaga-Vazquez MA, Chandler VL. Belele CL, et al. PLoS Genet. 2013;9(10):e1003773. doi: 10.1371/journal.pgen.1003773. Epub 2013 Oct 17. PLoS Genet. 2013. PMID: 24146624 Free PMC article. - Repeat elements and the Arabidopsis DNA methylation landscape.
Teixeira FK, Colot V. Teixeira FK, et al. Heredity (Edinb). 2010 Jul;105(1):14-23. doi: 10.1038/hdy.2010.52. Epub 2010 May 12. Heredity (Edinb). 2010. PMID: 20461104 Review. - Epigenetic silencing of endogenous repetitive sequences by MORPHEUS' MOLECULE1 in Arabidopsis thaliana.
Habu Y. Habu Y. Epigenetics. 2010 Oct 1;5(7):562-5. doi: 10.4161/epi.5.7.12518. Epigenetics. 2010. PMID: 21464619 Review.
Cited by
- Transposable elements: multifunctional players in the plant genome.
Hassan AH, Mokhtar MM, El Allali A. Hassan AH, et al. Front Plant Sci. 2024 Jan 4;14:1330127. doi: 10.3389/fpls.2023.1330127. eCollection 2023. Front Plant Sci. 2024. PMID: 38239225 Free PMC article. Review. - CRISPR-based targeting of DNA methylation in Arabidopsis thaliana by a bacterial CG-specific DNA methyltransferase.
Ghoshal B, Picard CL, Vong B, Feng S, Jacobsen SE. Ghoshal B, et al. Proc Natl Acad Sci U S A. 2021 Jun 8;118(23):e2125016118. doi: 10.1073/pnas.2125016118. Proc Natl Acad Sci U S A. 2021. PMID: 34074795 Free PMC article. - Genome-wide profiling and analysis of Arabidopsis siRNAs.
Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. Kasschau KD, et al. PLoS Biol. 2007 Mar;5(3):e57. doi: 10.1371/journal.pbio.0050057. PLoS Biol. 2007. PMID: 17298187 Free PMC article. - siRNA-mediated DNA methylation and H3K9 dimethylation in plants.
Xu C, Tian J, Mo B. Xu C, et al. Protein Cell. 2013 Sep;4(9):656-63. doi: 10.1007/s13238-013-3052-7. Epub 2013 Aug 13. Protein Cell. 2013. PMID: 23943321 Free PMC article. Review. - Signaling silence--breaking ground and spreading out.
Woo HR, Richards EJ. Woo HR, et al. Genes Dev. 2008 Jul 1;22(13):1719-23. doi: 10.1101/gad.1694608. Genes Dev. 2008. PMID: 18593872 Free PMC article.
References
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
- Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana . Nat Rev Genet. 2005;6:351–360. - PubMed
- Craig NL, Craigie RC, Gellert M, Lambowitz AM. Mobile DNA II. Washington (DC): ASM Press; 2002. 1182
- Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases