Proteolytic processing of delta-like 1 by ADAM proteases - PubMed (original) (raw)
Proteolytic processing of delta-like 1 by ADAM proteases
Emilia Dyczynska et al. J Biol Chem. 2007.
Abstract
Delta-like 1 (Dll1) is a mammalian ligand for Notch receptors. Interactions between Dll1 and Notch in trans activate the Notch pathway, whereas Dll1 binding to Notch in cis inhibits Notch signaling. Dll1 undergoes proteolytic processing in its extracellular domain by ADAM10. In this work we demonstrate that Dll1 represents a substrate for several other members of the ADAM family. In co-transfected cells, Dll1 is constitutively cleaved by ADAM12, and the N-terminal fragment of Dll1 is released to medium. ADAM12-mediated cleavage of Dll1 is cell density-dependent, takes place in cis orientation, and does not require the presence of the cytoplasmic domain of ADAM12. Full-length Dll1, but not its N- or C-terminal proteolytic fragment, co-immunoprecipitates with ADAM12. By using a Notch reporter construct, we show that Dll1 processing by ADAM12 increases Notch signaling in a cell-autonomous manner. Furthermore, ADAM9 and ADAM17 have the ability to process Dll1. In contrast, ADAM15 does not cleave Dll1, although the two proteins still co-immunoprecipitate with each other. Asn-353 present in the catalytic motif of ADAM12 and other Dll1-processing ADAMs, but absent in ADAM15, is necessary for Dll1 cleavage. Dll1 cleavage is reduced in ADAM9/12/15(-/-) mouse embryonic fibroblasts (MEFs), suggesting that the endogenous ADAM9 and/or ADAM12 present in wild type MEFs contribute to Dll1 processing. Finally, the endogenous Dll1 present in primary mouse myoblasts undergoes cleavage in confluent, differentiating myoblast cultures, and this cleavage is decreased by ADAM12 small interfering RNAs. Our findings expand the role of ADAM proteins in the regulation of Notch signaling.
Figures
FIG. 1. ADAM12 cleaves Dll1 in intact cells
A, COS-7 cells were transiently transfected with two empty vectors (lane 1), with mouse Delta-like 1 and empty ADAM12 expression vector (Dll1; lane 2), co-transfected with Dll1 and catalytically inactive mutant form of ADAM12 (E349Q A12; lane 3), or co-transfected with Dll1 and wild-type ADAM12 (A12; lane 4). Total cell lysates were analyzed by Western blotting using antibody specific for the C-terminus of Dll1 (top panel) or for the C-terminal domain of ADAM12 (middle panel). Full length (FL, 90-kDa band) and the C-terminal fragment (CTF, 29-kDa band) of Dll1 are indicated; the nascent pro-form of ADAM12 and the mature form of ADAM12 lacking the pro-domain are shown. The extent of Dll1 cleavage was determined as the ratio of intensities of the 29-kDa and 90-kDa Dll1 bands (mean ± S.E., n=5, bottom panel). B, Cells were transfected as in A, Dll1 construct contained c-Myc tag in the N-terminal domain. Cells were metabolically labeled with [35S]methionine+cysteine, cell lysates were analyzed by Western blotting using anti-c-Myc antibody (top) or anti-ADAM12 antibody (bottom), the N-terminal fragment (NTF) of Dll1 was immunoprecipitated from medium using anti-c-Myc antibody and visualized by SDS-PAGE and autoradiography (middle).
FIG. 2. Dll1 cleavage by ADAM12 is cell density-dependent and occurs in cis
A, COS-7 cells were co-transfected with Dll1 and either empty ADAM12 expression vector (lanes 1, 2) or with vector containing ADAM12 cDNA (lanes 3,4). Cells were ∼50% confluent (low density; L) or ∼90% confluent (high density; H) at the time of harvesting. B, COS-7 cells (∼90% confluent) were co-transfected with Dll1 and empty vector (lane 1) or Dll1 and ADAM12 (lane 2). Alternatively, cells (∼50% confluent) were transfected with Dll1 alone and co-cultured with vector-transfected (lane 3) or ADAM12-transfected cells (lane 4). In A and B, total cell lysates were analyzed by Western blotting using antibodies specific for the C-termini of Dll1 or ADAM12 (top and middle panels, respectively). The extent of Dll1 cleavage was determined as the ratio of intensities of the 29-kDa and 90-kDa Dll1 bands (mean ± S.E., n=3, bottom).
FIG. 3. ADAM12-mediated cleavage of Dll1 is followed by γ-secretase cleavage (A), is not stimulated by ionomycin, and is weakly enhanced by PMA (B)
A, COS-7 cells co-transfected with Dll1 and empty vector (lane 1) or ADAM12 (lanes 2-5) were incubated for 3 h with 5 μM lactacystin (a proteasomal inhibitor, lanes 3 and 5) or for 6 h with 1 μM L685,458 (a γ-secretase inhibitor, lanes 4 and 5). Cell lysates were analyzed by Western blotting using anti-Dll1 antibody. ICD, intracellular domain of Dll1, is the product of γ-secretase cleavage of CTF. B, COS-7 cells were co-transfected with Dll1 and either empty vector (lanes 1 and 2) or ADAM12 (lanes 3 and 4). Twenty four hours after transfection, cells were incubated for 5 h with 1 μM L685,458 and for additional 1 h with or without 5 μM ionomycin (left) or 25 ng/ml of PMA (right), as indicated. Cells were serum-starved for 30 min prior to and during the PMA treatment. Evidence for a role of ADAM17, but not ADAM12, in cleaving Notch1 (C). COS-7 cells were transfected with Notch1 containing C-terminal c-Myc tag (lanes 2-7, Notch-c-Myc) and either empty vector (lanes 1, 2, 5), ADAM12 (lanes 3 and 6), or ADAM17 (lanes 4 and 7). Two days after transfection, confluent cells were serum-starved for 30 min and then incubated for 1 h without (lanes 1-4) or with 25 ng/ml PMA (lanes 5-7), followed by analysis of Notch cleavage by Western blotting using anti-c-Myc antibody. TMIC, the S1 cleavage product; NEXT, Notch extracellular truncation, the S2 cleavage product; NICD, Notch intracellular domain, the S3 cleavage product.
FIG. 4. Cytoplasmic domain of ADAM12 is dispensable for Dll1 cleavage (A)
COS-7 cells were co-transfected with Dll1 and either the full length ADAM12 (lane 1) or ADAM12 containing a 169-amino acid deletion at the C-terminus (XT; lane 2). Total cell lysates were analyzed by Western blotting using antibody specific for the C-terminus of Dll1 (top) or the disintegrin domain of ADAM12 (bottom). The upper bands detected with anti-ADAM12 antibody in ADAM12- and XT-transfected cells represent the nascent forms of ADAM12 or XT, respectively, the lower bands represent the mature forms lacking the pro-domain. ADAM12 forms a complex with the full length Dll1, but not with NTF or CTF (B-D). B, Cells were co-transfected with Dll1 and ADAM12 (lanes 1 and 3) or with Dll1 and empty ADAM12 expression vector (lane 2), cell extracts were subjected to immunoprecipitation with antibody specific for the C-terminus of ADAM12 (lanes 1 and 2) or were incubated without antibody (lane 3), the immunoprecipitates (IPs) and inputs were analyzed by Western blotting using anti-Dll1 antibody. C, COS-7 cells were co-transfected with Dll1 and ADAM12, cell extracts were subjected to immunoprecipitation with (lane 1) or without (lane 2) anti-ADAM12 antibody; immunoprecipitates (IPs) and inputs were analyzed by immunoblotting with anti-Dll1 antibody. The secondary antibody was anti-rabbit IgG, Fc fragment-specific, and detected only the heavy chain of anti-ADAM12 antibody used for immunoprecipitation. Notice that the relative abundance of CTF Dll1 was much higher in the input than in the immunoprecipitate. D, Experiment was performed as in B using the Dll1 construct with c-Myc tag in the N-terminal domain. The immunoprecipitates (IPs) and inputs were analyzed by Western blotting using anti-c-Myc antibody.
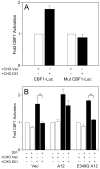
FIG. 5. Proteolytic processing of Dll1 by ADAM12 increases Notch signaling in a cell-autonomous manner
A, NIH3T3 cells were co-transfected with mouse Notch1 and either a CBF1 reporter (CBF1-Luc; pJH23A) or the same reporter in which CBF1 binding sites were mutated (Mut CBF1-Luc; pJH25A). After 24 h, cells were co-cultured with CHO cells stably transfected with vector only (CHO.Vec; white bars) or with CHO-cells stably transfected with Dll1 (CHO.Dll1; black bars). The activities of firefly luciferase, normalized to Renilla luciferase as internal control, were assayed 24 h later. B, NIH3T3 cells were transiently co-transfected with Notch1, a CBF1 reporter (pJT123A) and Dll1, wild type ADAM12 (A12), or catalytically inactive ADAM12 (E349Q A12), as indicated. After 24 h, cells were co-cultured for additional 24 h with CHO.Vec (white bars) or CHO.Dll1 cells (black bars), followed by measurement of luciferase activity. Notice that co-expression of Dll1 with Notch in the same cell inhibits Notch signaling induced by CHO.Dll1 (black bars) in the absence of A12 or in the presence of the E349Q mutant (*, p<0.05). In the presence of catalytically active A12, the inhibition was diminished and was not statistically significant. In A and B, error bars represent standard error of the mean (n=3).
FIG. 6. Dll1 is a substrate for ADAM9, 12, and 17, but not for ADAM15
A, COS-7 cells were co-transfected with Dll1 and empty vector (V), c-Myc-tagged ADAM9, 12, 15 or 17. Cell extracts were analyzed by Western blotting using anti-c-Myc tag antibody (top) or anti-Dll1 antibody (bottom). Upper bands detected with anti-c-Myc antibody represent nascent full length ADAM proteins, lower bands correspond to the catalytically active forms lacking pro-domains. B, COS-7 cells were co-transfected with Dll1 and empty vector (V), c-Myc-tagged ADAM9, 12, 15 or 17. ADAM proteins were immunoprecipitated with anti-c-Myc tag antibody. The inputs (left) and immunoprecipitates (IPs; right) were analyzed by anti-Dll1 (upper panels) and anti-c-Myc (lower panels) antibodies.
FIG. 7. Asn353 in ADAM12 is necessary for Dll1 cleavage
A, Alignment of the amino acid sequences of the catalytic sites of mouse ADAM9, 10, 12, 15, and 17. The consensus sequence of the catalytic site of ADAM proteases is shown on the top. B, COS-7 cells were co-transfected with Dll1 and either wild-type or N353S ADAM12 mutant, wild-type or S354N ADAM15 mutant, or empty vector. All ADAM constructs contained a C-terminal c-Myc tag. Cell extracts were analyzed by Western blotting using anti-Dll1 antibody (upper panel) or anti-c-Myc antibody (bottom panel). C, COS-7 cells were co-transfected with Dll1 and wild-type ADAM12, the N353S mutant of ADAM12, or empty vector, as indicated. Cell extracts were immunoprecipitated using anti-ADAM12 antibody (lanes 1-3) or were incubated without antibody (lane 4). Immunoprecipitates (IPs) and inputs were analyzed by Western blotting with anti-Dll1 antibody, the expression of ADAM12 was verified by Western blotting of the inputs with anti-ADAM12 antibody. D, The predicted structure of the metalloprotease domain of ADAM12, obtained by threading mouse ADAM12 sequence (amino acids 212-414) onto mouse ADAM33 (PDB structure 1R55) using the Swiss-PdbViewer software, and then submitted to the SWISS-MODEL server for refinement. Side chains of His348, His352, and His358 coordinating Zn2+ are shown in blue, catalytic E349 is shown in red, and N353 is depicted in green.
FIG. 8. Endogenous ADAM9 and/or ADAM12 contribute to Dll1 cleavage in mouse embryonic fibroblasts (MEFs)
SV40-immortalized MEFs isolated from wild type mice (wt) or from ADAM9/12/15−/− triple knockout mice (T) were transiently transfected to express Dll1, as indicated. A, After 48 h, Dll1 processing was analyzed by Western blotting with anti-Dll1 antibody. The intensities of the bands corresponding to FL Dll1 were determined after short exposure times (left), the intensities of the bands representing CTF Dll1 were measured after long exposure times (right). The amounts of FL Dll1 and CTF Dll1 in T-MEFs were normalized to the amounts in wt-MEFs; the results (mean from three different experiments, ± S.E) are shown below each Western blot. B, Twenty four hours after transfection, cells were metabolically labeled with [35S]methionine+cysteine, 16 h later cell lysates were subjected to immunoprecipitation using anti-Dll1 antibody, as indicated. The immunoprecipitates were resolved by SDS-PAGE and analyzed by autoradiography. The intensities of the bands corresponding to FL Dll1 and CTF Dll1 were determined after short exposure times (left) and long exposure times (right), respectively, and the results were plotted as in A (mean ± S.E., n=2). In A and B, cells were incubated for 6 h in the presence of 1 μM γ-secretase inhibitor L685,458 prior to harvesting.
FIG. 9. Endogenous ADAM12 co-immunoprecipitates with Dll1 and contributes to Dll1 cleavage in primary mouse myoblasts
A, Primary mouse myoblasts grown to confluence and incubated for one day in differentiation medium were subjected to immunoprecipitation with anti-ADAM12 antibody. The immunoprecipitates were analyzed by immunoblotting with anti-ADAM12 (top) or anti-Dll1 antibody (bottom). B, Primary mouse myoblasts were transfected with one of three ADAM12 siRNAs or control siRNA, as described in Experimental Procedures. One day after transfection, cells reached confluence and were transferred to differentiation medium. After 24 h, cells were metabolically labeled with [35S]methionine+cysteine for 10 h, 1 μM γ-secretase inhibitor L685,458 was added, and the labeling continued for next 6 h. The level of endogenous ADAM12 was analyzed by Western blotting after partial purification on concanavalin A agarose (bottom). In parallel, the endogenous Dll1 was immunoprecipitated with anti-Dll1 antibody, the immunoprecipitates were analyzed by SDS-PAGE and direct autoradiography (top). The exposure time of the region in the autoradiogram containing the FL Dll1 was 5 times longer that the exposure time of the region containing CTF Dll1. Notice that the abundance of CTF Dll1 is diminished in cells transfected with ADAM12 siRNAs.
Similar articles
- The role of Delta-like 1 shedding in muscle cell self-renewal and differentiation.
Sun D, Li H, Zolkiewska A. Sun D, et al. J Cell Sci. 2008 Nov 15;121(Pt 22):3815-23. doi: 10.1242/jcs.035493. Epub 2008 Oct 28. J Cell Sci. 2008. PMID: 18957511 Free PMC article. - Inhibition of proteolysis of Delta-like-1 does not promote or reduce T-cell developmental potential.
Gravano DM, Manilay JO. Gravano DM, et al. Immunol Cell Biol. 2010 Oct;88(7):746-53. doi: 10.1038/icb.2010.30. Epub 2010 Mar 16. Immunol Cell Biol. 2010. PMID: 20231851 - Metalloprotease-disintegrin ADAM12 expression is regulated by Notch signaling via microRNA-29.
Li H, Solomon E, Duhachek Muggy S, Sun D, Zolkiewska A. Li H, et al. J Biol Chem. 2011 Jun 17;286(24):21500-10. doi: 10.1074/jbc.M110.207951. Epub 2011 Apr 25. J Biol Chem. 2011. PMID: 21518768 Free PMC article. - ADAM proteases: ligand processing and modulation of the Notch pathway.
Zolkiewska A. Zolkiewska A. Cell Mol Life Sci. 2008 Jul;65(13):2056-68. doi: 10.1007/s00018-008-7586-4. Cell Mol Life Sci. 2008. PMID: 18344021 Free PMC article. Review. - Cellular roles of ADAM12 in health and disease.
Kveiborg M, Albrechtsen R, Couchman JR, Wewer UM. Kveiborg M, et al. Int J Biochem Cell Biol. 2008;40(9):1685-702. doi: 10.1016/j.biocel.2008.01.025. Epub 2008 Feb 1. Int J Biochem Cell Biol. 2008. PMID: 18342566 Review.
Cited by
- Identification of ILK as a new partner of the ADAM12 disintegrin and metalloprotease in cell adhesion and survival.
Leyme A, Bourd-Boittin K, Bonnier D, Falconer A, Arlot-Bonnemains Y, Théret N. Leyme A, et al. Mol Biol Cell. 2012 Sep;23(17):3461-72. doi: 10.1091/mbc.E11-11-0918. Epub 2012 Jul 5. Mol Biol Cell. 2012. PMID: 22767580 Free PMC article. - Turn It Down a Notch.
Carrieri FA, Dale JK. Carrieri FA, et al. Front Cell Dev Biol. 2017 Jan 18;4:151. doi: 10.3389/fcell.2016.00151. eCollection 2016. Front Cell Dev Biol. 2017. PMID: 28149836 Free PMC article. Review. - Control of endothelial cell tube formation by Notch ligand intracellular domain interactions with activator protein 1 (AP-1).
Forghany Z, Robertson F, Lundby A, Olsen JV, Baker DA. Forghany Z, et al. J Biol Chem. 2018 Jan 26;293(4):1229-1242. doi: 10.1074/jbc.M117.819045. Epub 2017 Dec 1. J Biol Chem. 2018. PMID: 29196606 Free PMC article. - Hypoxia-induced release, nuclear translocation, and signaling activity of a DLK1 intracellular fragment in glioma.
Grassi ES, Pantazopoulou V, Pietras A. Grassi ES, et al. Oncogene. 2020 May;39(20):4028-4044. doi: 10.1038/s41388-020-1273-9. Epub 2020 Mar 24. Oncogene. 2020. PMID: 32205867 Free PMC article. - The role of Adams in Notch signaling.
Groot AJ, Vooijs MA. Groot AJ, et al. Adv Exp Med Biol. 2012;727:15-36. doi: 10.1007/978-1-4614-0899-4_2. Adv Exp Med Biol. 2012. PMID: 22399336 Free PMC article. Review.
References
- Lai EC. Development. 2004;131:965–973. - PubMed
- Kadesch T. Curr. Opin. Genet. Dev. 2004;14:506–512. - PubMed
- Louvi A, Artavanis-Tsakonas S. Nat. Rev. Neurosci. 2006;7:93–102. - PubMed
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israël A. Mol. Cell. 2000;5:207–216. - PubMed
- Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. Mol. Cell. 2000;5:197–206. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM065528/GM/NIGMS NIH HHS/United States
- R01 GM064750/GM/NIGMS NIH HHS/United States
- P20 RR017708/RR/NCRR NIH HHS/United States
- R01 GM065528/GM/NIGMS NIH HHS/United States
- GM64750/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous