Structure of the catalytic pore of gamma-secretase probed by the accessibility of substituted cysteines - PubMed (original) (raw)
Structure of the catalytic pore of gamma-secretase probed by the accessibility of substituted cysteines
Chihiro Sato et al. J Neurosci. 2006.
Abstract
Several single-span membrane proteins are cleaved within their transmembrane domains (TMDs) by intramembrane-cleaving proteases, although the structure of the active site executing intramembrane cleavage remains unknown. Here we use the substituted cysteine accessibility method to examine the structure of presenilin-1, a catalytic subunit of gamma-secretase, involved in amyloid beta protein generation in Alzheimer's disease and Notch signaling. We show that TMD6 and TMD7 of presenilin-1 contribute to the formation of a hydrophilic pore within the membrane. Residues at the luminal portion of TMD6 are predicted to form a subsite for substrate or inhibitor binding on the alpha-helix facing a hydrophilic milieu, whereas those around the GxGD catalytic motif within TMD7 are highly water accessible, suggesting formation of a hydrophilic structure within the pore. Collectively, our data suggest that the active site of gamma-secretase resides in a catalytic pore filled with water within the lipid bilayer and is tapered around the catalytic aspartates.
Figures
Figure 1.
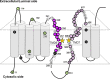
Locations of the PS1 cysteine mutations used in this study. A schematic depiction of human PS1 based on a 9 TMD topology is shown. Catalytic aspartates are shown by yellow stars. Endogenous cysteines replaced with serine in PS1/Cys(−) are indicated by black circles. Amino acid residues substituted to cysteines are shown by a circle with a single-letter character representing the original amino acid. Mutated cysteines that retained γ-secretase activity and thus were analyzed as controls are indicated as green circles. Single-Cys mt PS1 in HR6 and HR8 analyzed in this study are indicated by purple and pink circles, respectively. Cysteine substitutions that resulted in a loss of γ-secretase activity are indicated by gray circles.
Figure 2.
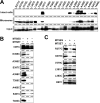
SCAM analysis of single-Cys mt PS1 at representative positions. A, Western blot analysis of single-Cys mt PS1 stably expressed in DKO cells. Endoproteolysis of PS1 and restoration of the expression of mature glycosylated Nct and Pen-2 in all single-Cys mt PS1 are shown. B, Effect of single-Cys mt PS1 on Aβ levels secreted from DKO cells coexpressing APPNL. Error bars indicate mean ± SE (n = 3). C, Biotin-labeling experiment using MTSEA-biotin in intact cells (left) and microsomes (right). D, Labeling competition by preincubation with MTSES and MTSET. MEF, Mouse embryonic fibroblast; wt, wild type.
Figure 3.
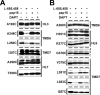
SCAM analysis of single-Cys mt PS1 in HR6. A, Biotin-labeling experiment using MTSEA-biotin in intact cells (top) and microsomes (middle). The amount of PS1 NTF in the input fraction is shown in the bottom panel. B, Labeling competition by MTSES and MTSET.
Figure 4.
SCAM analysis of single-Cys mt PS1 around HR8. A, Biotin-labeling experiment using MTSEA-biotin in intact cells (top) and microsomes (middle). The amount of PS1 CTF in the input fraction is shown in the bottom panel. B, C, Biotin labeling of single-Cys mt PS1 around HR8 was performed after preincubation with MTSES or MTSET in intact cells (B) or microsomes (C). Locations of cysteine mutations are shown on the left side of the panel.
Figure 5.
Labeling competition by γ-secretase inhibitors. Biotin labeling of single-Cys mt PS1 was conducted after preincubation with L-685,458, pep15, or DAPT in intact cells (A) or microsomes (B). Locations and predicted topology of cysteine mutations are shown on the left and right sides of the panel, respectively.
Figure 6.
Cross-linking experiment using MTS cross-linkers. Thiol-mediated cross-linking experiments of single-Cys or double-Cys mt in TMD6 (L250C) and TMD7 (L383C and I387C) using MTS cross-linkers are shown.
Figure 7.
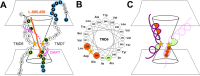
Hypothetical structure around the catalytic site of γ-secretase. A, A summary of SCAM analysis. Catalytic aspartates are indicated by yellow stars. Cysteine mutants that were labeled by MTSEA-biotin are shown by a white letter in a black circle. The labeling of residues that were effectively competed by MTSES or MTSET is shown in a blue frame, and the unaffected residues are in a green frame. Residues that were not labeled by MTSEA-biotin or unanalyzed are shown by a black letter in a white and gray circle, respectively. Putative locations of L-685,458 and DAPT within the catalytic pore of PS1 are indicated by orange and pink lines, respectively. B, α-Helical wheel alignment consisting of the 18 amino acids of TMD6 of human PS1 starting at the N-terminal tryptophan 244. Catalytic aspartate at position 257 is indicated by a star. Residues that were biotin labeled and competed by L-685,458 (i.e., A246 and L250) are shown by orange circles. A260 was indicated by a green circle. Note that A246 and L250 are aligned on the same interface with D257 in an α-helical model. C, Schematic depiction of the configuration of the catalytic pore. TMD6 and TMD7 are shown in purple and pink lines, respectively. Catalytic aspartates are shown in yellow stars. Regions that serve as subsites, including the conserved motif forming a catalytic site (pink oval), are indicated in orange. Residues that are close to the aspartates but putatively facing a geometrically different hydrophilic environment from the catalytic pore are shown in green.
Similar articles
- The C-terminal PAL motif and transmembrane domain 9 of presenilin 1 are involved in the formation of the catalytic pore of the gamma-secretase.
Sato C, Takagi S, Tomita T, Iwatsubo T. Sato C, et al. J Neurosci. 2008 Jun 11;28(24):6264-71. doi: 10.1523/JNEUROSCI.1163-08.2008. J Neurosci. 2008. PMID: 18550769 Free PMC article. - Participation of transmembrane domain 1 of presenilin 1 in the catalytic pore structure of the γ-secretase.
Takagi S, Tominaga A, Sato C, Tomita T, Iwatsubo T. Takagi S, et al. J Neurosci. 2010 Nov 24;30(47):15943-50. doi: 10.1523/JNEUROSCI.3318-10.2010. J Neurosci. 2010. PMID: 21106832 Free PMC article. - Conformational Changes in Transmembrane Domain 4 of Presenilin 1 Are Associated with Altered Amyloid-β 42 Production.
Tominaga A, Cai T, Takagi-Niidome S, Iwatsubo T, Tomita T. Tominaga A, et al. J Neurosci. 2016 Jan 27;36(4):1362-72. doi: 10.1523/JNEUROSCI.5090-14.2016. J Neurosci. 2016. PMID: 26818522 Free PMC article. - Molecular mechanism of intramembrane proteolysis by γ-secretase.
Tomita T. Tomita T. J Biochem. 2014 Oct;156(4):195-201. doi: 10.1093/jb/mvu049. Epub 2014 Aug 9. J Biochem. 2014. PMID: 25108625 Review. - The Alzheimer's disease-associated gamma-secretase complex: functional domains in the presenilin 1 protein.
Laudon H, Winblad B, Näslund J. Laudon H, et al. Physiol Behav. 2007 Sep 10;92(1-2):115-20. doi: 10.1016/j.physbeh.2007.05.037. Epub 2007 May 21. Physiol Behav. 2007. PMID: 17588625 Review.
Cited by
- Glutamate Transporter 1 as a Novel Negative Regulator of Amyloid β.
Sinha P, Turchyna Y, Mitchell SPC, Sadek M, Armagan G, Perrin F, Maesako M, Berezovska O. Sinha P, et al. Cells. 2024 Sep 24;13(19):1600. doi: 10.3390/cells13191600. Cells. 2024. PMID: 39404364 Free PMC article. - Presenilin-like GxGD membrane proteases have dual roles as proteolytic enzymes and ion channels.
Kuo IY, Hu J, Ha Y, Ehrlich BE. Kuo IY, et al. J Biol Chem. 2015 Mar 6;290(10):6419-27. doi: 10.1074/jbc.M114.629584. Epub 2015 Jan 21. J Biol Chem. 2015. PMID: 25609250 Free PMC article. - Modulating Hinge Flexibility in the APP Transmembrane Domain Alters γ-Secretase Cleavage.
Götz A, Mylonas N, Högel P, Silber M, Heinel H, Menig S, Vogel A, Feyrer H, Huster D, Luy B, Langosch D, Scharnagl C, Muhle-Goll C, Kamp F, Steiner H. Götz A, et al. Biophys J. 2019 Jun 4;116(11):2103-2120. doi: 10.1016/j.bpj.2019.04.030. Epub 2019 May 3. Biophys J. 2019. PMID: 31130234 Free PMC article. - Nicastrin is dispensable for gamma-secretase protease activity in the presence of specific presenilin mutations.
Futai E, Yagishita S, Ishiura S. Futai E, et al. J Biol Chem. 2009 May 8;284(19):13013-22. doi: 10.1074/jbc.M807653200. Epub 2009 Mar 2. J Biol Chem. 2009. PMID: 19254953 Free PMC article. - Contribution of the γ-secretase subunits to the formation of catalytic pore of presenilin 1 protein.
Takeo K, Watanabe N, Tomita T, Iwatsubo T. Takeo K, et al. J Biol Chem. 2012 Jul 27;287(31):25834-43. doi: 10.1074/jbc.M111.336347. Epub 2012 Jun 11. J Biol Chem. 2012. PMID: 22689582 Free PMC article.
References
- Akabas MH, Kaufmann C, Cook TA, Archdeacon P. Amino acid residues lining the chloride channel of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1994;269:14865–14868. - PubMed
- Cervantes S, Saura CA, Pomares E, Gonzalez-Duarte R, Marfany G. Functional implications of the presenilin dimerization: reconstitution of γ-secretase activity by assembly of a catalytic site at the dimer interface of two catalytically inactive presenilins. J Biol Chem. 2004;279:36519–36529. - PubMed
- Das C, Berezovska O, Diehl TS, Genet C, Buldyrev I, Tsai JY, Hyman BT, Wolfe MS. Designed helical peptides inhibit an intramembrane protease. J Am Chem Soc. 2003;125:11794–11795. - PubMed
- Doan A, Thinakaran G, Borchelt DR, Slunt HH, Ratovitsky T, Podlisny M, Selkoe DJ, Seeger M, Gandy SE, Price DL, Sisodia SS. Protein topology of presenilin 1. Neuron. 1996;17:1023–1030. - PubMed
- Esler WP, Kimberly WT, Ostaszewski BL, Diehl TS, Moore CL, Tsai JY, Rahmati T, Xia W, Selkoe DJ, Wolfe MS. Transition-state analogue inhibitors of γ-secretase bind directly to presenilin-1. Nat Cell Biol. 2000;2:428–434. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources