miRGen: a database for the study of animal microRNA genomic organization and function - PubMed (original) (raw)
. 2007 Jan;35(Database issue):D149-55.
doi: 10.1093/nar/gkl904. Epub 2006 Nov 15.
Affiliations
- PMID: 17108354
- PMCID: PMC1669779
- DOI: 10.1093/nar/gkl904
miRGen: a database for the study of animal microRNA genomic organization and function
Molly Megraw et al. Nucleic Acids Res. 2007 Jan.
Abstract
miRGen is an integrated database of (i) positional relationships between animal miRNAs and genomic annotation sets and (ii) animal miRNA targets according to combinations of widely used target prediction programs. A major goal of the database is the study of the relationship between miRNA genomic organization and miRNA function. This is made possible by three integrated and user friendly interfaces. The Genomics interface allows the user to explore where whole-genome collections of miRNAs are located with respect to UCSC genome browser annotation sets such as Known Genes, Refseq Genes, Genscan predicted genes, CpG islands and pseudogenes. These miRNAs are connected through the Targets interface to their experimentally supported target genes from TarBase, as well as computationally predicted target genes from optimized intersections and unions of several widely used mammalian target prediction programs. Finally, the Clusters interface provides predicted miRNA clusters at any given inter-miRNA distance and provides specific functional information on the targets of miRNAs within each cluster. All of these unique features of miRGen are designed to facilitate investigations into miRNA genomic organization, co-transcription and targeting. miRGen can be freely accessed at http://www.diana.pcbi.upenn.edu/miRGen.
Figures
Figure 1
miRGen Genomics interface links to the UCSC genome browser and to the miRGen Targets interface Links to the UCSC genome browser for each miRNA and gene provide graphical views. Links to the miRGen Targets interface provide tabular views of predicted or experimentally supported targets.
Figure 2
miRGen Clusters Interface Users can choose which interface that the clusters should link to. The figure above shows links to the ‘Predicted Targets Optimized Intersection (PicTar, TargetScanS)’ target set and to the UCSC view for a particular cluster.
Figure 3
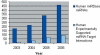
Growth in Number of miRNAs and Experimentally Supported Targets This chart tracks the number of Human miRNAs in miRBase and the number of experimentally supported target instances, published through recent years.
Similar articles
- miRNAMap: genomic maps of microRNA genes and their target genes in mammalian genomes.
Hsu PW, Huang HD, Hsu SD, Lin LZ, Tsou AP, Tseng CP, Stadler PF, Washietl S, Hofacker IL. Hsu PW, et al. Nucleic Acids Res. 2006 Jan 1;34(Database issue):D135-9. doi: 10.1093/nar/gkj135. Nucleic Acids Res. 2006. PMID: 16381831 Free PMC article. - miRGen 2.0: a database of microRNA genomic information and regulation.
Alexiou P, Vergoulis T, Gleditzsch M, Prekas G, Dalamagas T, Megraw M, Grosse I, Sellis T, Hatzigeorgiou AG. Alexiou P, et al. Nucleic Acids Res. 2010 Jan;38(Database issue):D137-41. doi: 10.1093/nar/gkp888. Epub 2009 Oct 22. Nucleic Acids Res. 2010. PMID: 19850714 Free PMC article. - miRBase: the microRNA sequence database.
Griffiths-Jones S. Griffiths-Jones S. Methods Mol Biol. 2006;342:129-38. doi: 10.1385/1-59745-123-1:129. Methods Mol Biol. 2006. PMID: 16957372 Review. - ViTa: prediction of host microRNAs targets on viruses.
Hsu PW, Lin LZ, Hsu SD, Hsu JB, Huang HD. Hsu PW, et al. Nucleic Acids Res. 2007 Jan;35(Database issue):D381-5. doi: 10.1093/nar/gkl1009. Epub 2006 Dec 5. Nucleic Acids Res. 2007. PMID: 17148483 Free PMC article. - Computational prediction of microRNA genes.
Hertel J, Langenberger D, Stadler PF. Hertel J, et al. Methods Mol Biol. 2014;1097:437-56. doi: 10.1007/978-1-62703-709-9_20. Methods Mol Biol. 2014. PMID: 24639171 Review.
Cited by
- MiR-429 inhibits oral squamous cell carcinoma growth by targeting ZEB1.
Lei W, Liu YE, Zheng Y, Qu L. Lei W, et al. Med Sci Monit. 2015 Feb 2;21:383-9. doi: 10.12659/MSM.893412. Med Sci Monit. 2015. PMID: 25640197 Free PMC article. - Expression analysis in multiple muscle groups and serum reveals complexity in the microRNA transcriptome of the mdx mouse with implications for therapy.
Roberts TC, Blomberg KE, McClorey G, El Andaloussi S, Godfrey C, Betts C, Coursindel T, Gait MJ, Smith CI, Wood MJ. Roberts TC, et al. Mol Ther Nucleic Acids. 2012 Aug 14;1(8):e39. doi: 10.1038/mtna.2012.26. Mol Ther Nucleic Acids. 2012. PMID: 23344181 Free PMC article. - ChemiRs: a web application for microRNAs and chemicals.
Su EC, Chen YS, Tien YC, Liu J, Ho BC, Yu SL, Singh S. Su EC, et al. BMC Bioinformatics. 2016 Apr 18;17:167. doi: 10.1186/s12859-016-1002-0. BMC Bioinformatics. 2016. PMID: 27091357 Free PMC article. - MicroRNAs in autophagy and their emerging roles in crosstalk with apoptosis.
Xu J, Wang Y, Tan X, Jing H. Xu J, et al. Autophagy. 2012 Jun;8(6):873-82. doi: 10.4161/auto.19629. Epub 2012 Mar 23. Autophagy. 2012. PMID: 22441107 Free PMC article. Review. - Assessment of mRNA and microRNA Stabilization in Peripheral Human Blood for Multicenter Studies and Biobanks.
Weber DG, Casjens S, Rozynek P, Lehnert M, Zilch-Schöneweis S, Bryk O, Taeger D, Gomolka M, Kreuzer M, Otten H, Pesch B, Johnen G, Brüning T. Weber DG, et al. Biomark Insights. 2010 Sep 22;5:95-102. doi: 10.4137/bmi.s5522. Biomark Insights. 2010. PMID: 20981139 Free PMC article.
References
- Lee R.C., Feinbaum R.L., Ambros V. The C.elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. - PubMed
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. - PubMed
- Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. - PubMed
- Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources