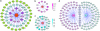NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth - PubMed (original) (raw)
. 2006 Nov 28;103(48):18261-6.
doi: 10.1073/pnas.0606108103. Epub 2006 Nov 17.
Wei Keat Lim, Duncan T Odom, Maria Luisa Sulis, Pedro J Real, Adam Margolin, Kelly C Barnes, Jennifer O'Neil, Donna Neuberg, Andrew P Weng, Jon C Aster, Francois Sigaux, Jean Soulier, A Thomas Look, Richard A Young, Andrea Califano, Adolfo A Ferrando
Affiliations
- PMID: 17114293
- PMCID: PMC1838740
- DOI: 10.1073/pnas.0606108103
NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth
Teresa Palomero et al. Proc Natl Acad Sci U S A. 2006.
Erratum in
- Proc Natl Acad Sci U S A. 2007 Mar 6;104(10):4240
Abstract
The NOTCH1 signaling pathway directly links extracellular signals with transcriptional responses in the cell nucleus and plays a critical role during T cell development and in the pathogenesis over 50% of human T cell lymphoblastic leukemia (T-ALL) cases. However, little is known about the transcriptional programs activated by NOTCH1. Using an integrative systems biology approach we show that NOTCH1 controls a feed-forward-loop transcriptional network that promotes cell growth. Inhibition of NOTCH1 signaling in T-ALL cells led to a reduction in cell size and elicited a gene expression signature dominated by down-regulated biosynthetic pathway genes. By integrating gene expression array and ChIP-on-chip data, we show that NOTCH1 directly activates multiple biosynthetic routes and induces c-MYC gene expression. Reverse engineering of regulatory networks from expression profiles showed that NOTCH1 and c-MYC govern two directly interconnected transcriptional programs containing common target genes that together regulate the growth of primary T-ALL cells. These results identify c-MYC as an essential mediator of NOTCH1 signaling and integrate NOTCH1 activation with oncogenic signaling pathways upstream of c-MYC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Oncogenic NOTCH1 signaling activates transcriptional programs regulating leukemic cell growth. (a) Activated intracellular NOTCH1 levels after γ-secretase cleavage were detected by Western blot analysis using the Val-1744 antibody that specifically recognizes ICN1, in cell lysates from T-ALL cell lines treated with GSI (CompE 500 nM) for 24 h or mock-treated (DMSO) controls. Smaller molecular weight bands (ICN1-ΔPEST) are observed in cell lines with truncating mutations in the PEST domain of NOTCH1. α-Tubulin levels are shown as loading control. (b) Gene expression profiling was performed in duplicate samples from 7 T-ALL cell lines treated with GSI (CompE) or vehicle (DMSO) for 24 h. Heat map represents color coded expression levels for each sample with respect to mock treatment controls. Top significant genes (P < 0.00001) ranked by t test are shown. (c) Functional annotation of top genes (P < 0.0001) down-regulated after NOTCH inhibition by GSI treatment using the DAVID tool. (d) Detailed heat maps representing expression changes induced by GSI in selected functional categories.
Fig. 2.
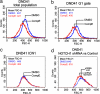
Inhibition of NOTCH1 signaling impairs cell growth in T-ALL cells. (a) DND41 T-ALL cells treated with GSI for 6 days showed a reduction in cell diameters compared with vehicle (DMSO) treatment. Representative histograms of triplicate experiments are shown. Similar results were obtained in HPB-ALL, CUTLL1, and KOPTK1 cell lines. (b) GSI-induced changes in cell size are independent of cell cycle as demonstrated by analysis of G1 (2N DNA content) gated cells. (c) Forced expression of ICN1 rescues the effects of GSI in cell size. (d) Inhibition of NOTCH1 signaling via lentiviral expression of shRNAs targeting NOTCH1 (pGK GFP shRNA NOTCH1) or the luciferase gene (pGK GFP shRNA LUC) used as control.
Fig. 3.
NOTCH1 is a direct regulator of genes involved in cell growth and metabolism. (a) Scatter plot showing the combined results of three independent ChIP-on-chip experiments with the N1-TAD antibody recognizing the transactivation domain of NOTCH1 in HPB-ALL cells. The log fluorescence values for control DNA labeled with Cy3 are plotted on the x axis, and those for the NOTCH1 IP DNA labeled with Cy5 are plotted on the y axis. Promoter sequences bound by NOTCH1 (arrowheads) are shifted to the upper left side from the diagonal. (b) Functional classification of NOTCH1 direct targets identified by ChIP-on-chip (P < 0.0001). (c) Top gene expression changes induced by GSI in T-ALL cell lines ranked by signal-to-noise ratio. Heat map represents color-coded expression levels for each sample with respect to mock treatment controls. Previously known NOTCH1 direct target genes and genes identified on ChIP-on-chip analysis are highlighted. (d) Graphic representation of the GSEA enrichment score and distribution of NOTCH1 occupied promoter genes (binding P value <0.0001) along the rank of GSI-regulated transcripts. (e) Annotation of NOTCH1 direct target genes along the top GSI-regulated transcripts ranked by signal-to-noise ratio.
Fig. 4.
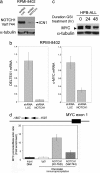
NOTCH1 binds to c-MYC proximal promoter sequences and regulates c-MYC mRNA and protein expression in T-ALL. (a) Western blot analysis of ICN1 levels in RPMI 8402 T-ALL cells infected with lentiviruses (pLKO puro) driving expression of shRNAs targeting NOTCH1 (shRNA NOTCH1) or control shRNAs targeting the luciferase gene (shRNA LUC). (b) Quantitative RT-PCR analysis of DELTEX1 and c-MYC transcript levels in RPMI 8402 T-ALL cells upon NOTCH1 shRNA knockdown and in control shRNA-treated cells. (c) Western blot analysis of c-MYC in HPB-ALL T-ALL cells showing down-regulation of c-MYC expression upon NOTCH1 inhibition with GSI (CompE 500 nM). The arrowhead indicates the c-MYC-specific band. (d) ChIP analysis of NOTCH1 binding to c-MYC promoter sequences. Quantitative PCR analysis of c-MYC promoter sequences normalized to β-actin levels in control DNA and chromatin immunoprecipitates performed with two NOTCH1 antibodies (N1-TAD and Val 1744) and IgG as control in HPB-ALL cells.
Fig. 5.
NOTCH and c-MYC constitute a feed-forward regulatory network motif controlling cell growth. (a) Structure of the NOTCH1 feed-forward-loop regulatory motif controlling c-MYC and cell growth genes. (b) NOTCH1 inhibition with GSI in T-ALL cells switches off genes up-regulated by forced expression of c-MYC in T cell lymphoblasts. Graphic representation of the GSEA enrichment score and distribution of c-MYC-regulated genes along the rank of GSI-regulated transcripts. (c) Retroviral expression of c-MYC in DND41 rescues cell growth after GSI treatment. Cell size was estimated by flow cytometry in DND41 cells infected with pBMN c-MYC IRES GFP retrovirus growing in the presence of CompE or vehicle (DMSO) for 4 days.
Fig. 6.
Activated NOTCH1 and c-MYC regulatory network in T-ALL. (a) A metagene based on the gene expression signature associated with levels of activated NOTCH1 protein in T-ALL cell lines was integrated in an ARACNe global regulatory network constructed with microarray expression data from T-ALL samples. Neighbors of the NOTCH1 metagene identified as NOTCH1 direct target genes by ChIP-on-chip are shaded in pink, neighbors regulated in T-ALL cells treated with a GSI are shown shaded in blue, and neighbor genes showing both significant promoter occupancy by ChIP-on-chip and regulation upon GSI treatment are shaded in purple. (b) Detailed representation of overlap between NOTCH1 neighbor genes and ChIP-on-chip data. The intensity of each neighboring node represents the significance level of promoter occupancy by NOTCH1. (c) Detailed representation of NOTCH1 neighbor genes regulated in T-ALL cells treated with a GSI. The intensity of each neighboring node, corresponding to the scale panel on the right, represents the significance level of gene regulation by GSI treatment. (d) Representation of direct neighbor genes associated with the NOTCH1 metagene and with c-MYC.
Similar articles
- Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc.
Sharma VM, Calvo JA, Draheim KM, Cunningham LA, Hermance N, Beverly L, Krishnamoorthy V, Bhasin M, Capobianco AJ, Kelliher MA. Sharma VM, et al. Mol Cell Biol. 2006 Nov;26(21):8022-31. doi: 10.1128/MCB.01091-06. Epub 2006 Sep 5. Mol Cell Biol. 2006. PMID: 16954387 Free PMC article. - The NOTCH1-MYC highway toward T-cell acute lymphoblastic leukemia.
Sanchez-Martin M, Ferrando A. Sanchez-Martin M, et al. Blood. 2017 Mar 2;129(9):1124-1133. doi: 10.1182/blood-2016-09-692582. Epub 2017 Jan 23. Blood. 2017. PMID: 28115368 Review. - Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice.
Klinakis A, Szabolcs M, Politi K, Kiaris H, Artavanis-Tsakonas S, Efstratiadis A. Klinakis A, et al. Proc Natl Acad Sci U S A. 2006 Jun 13;103(24):9262-7. doi: 10.1073/pnas.0603371103. Epub 2006 Jun 2. Proc Natl Acad Sci U S A. 2006. PMID: 16751266 Free PMC article. - c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma.
Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, Li Y, Wolfe MS, Shachaf C, Felsher D, Blacklow SC, Pear WS, Aster JC. Weng AP, et al. Genes Dev. 2006 Aug 1;20(15):2096-109. doi: 10.1101/gad.1450406. Epub 2006 Jul 17. Genes Dev. 2006. PMID: 16847353 Free PMC article. - Oncogenic NOTCH1 control of MYC and PI3K: challenges and opportunities for anti-NOTCH1 therapy in T-cell acute lymphoblastic leukemias and lymphomas.
Palomero T, Ferrando A. Palomero T, et al. Clin Cancer Res. 2008 Sep 1;14(17):5314-7. doi: 10.1158/1078-0432.CCR-07-4864. Clin Cancer Res. 2008. PMID: 18765521 Free PMC article. Review.
Cited by
- Determinants and role of chromatin organization in acute leukemia.
Fang C, Rao S, Crispino JD, Ntziachristos P. Fang C, et al. Leukemia. 2020 Oct;34(10):2561-2575. doi: 10.1038/s41375-020-0981-z. Epub 2020 Jul 20. Leukemia. 2020. PMID: 32690881 Free PMC article. Review. - NOTCH1 mutations influence survival in chronic lymphocytic leukemia patients.
Willander K, Dutta RK, Ungerbäck J, Gunnarsson R, Juliusson G, Fredrikson M, Linderholm M, Söderkvist P. Willander K, et al. BMC Cancer. 2013 Jun 4;13:274. doi: 10.1186/1471-2407-13-274. BMC Cancer. 2013. PMID: 23734977 Free PMC article. - HES1 opposes a PTEN-dependent check on survival, differentiation, and proliferation of TCRβ-selected mouse thymocytes.
Wong GW, Knowles GC, Mak TW, Ferrando AA, Zúñiga-Pflücker JC. Wong GW, et al. Blood. 2012 Aug 16;120(7):1439-48. doi: 10.1182/blood-2011-12-395319. Epub 2012 May 30. Blood. 2012. PMID: 22649105 Free PMC article. - An oncogenic enhancer enemy (N-Me) in T-ALL.
Herranz D, Ferrando AA. Herranz D, et al. Cell Cycle. 2015;14(2):167-8. doi: 10.4161/15384101.2014.989129. Cell Cycle. 2015. PMID: 25584678 Free PMC article. No abstract available. - Genomics in acute lymphoblastic leukaemia: insights and treatment implications.
Roberts KG, Mullighan CG. Roberts KG, et al. Nat Rev Clin Oncol. 2015 Jun;12(6):344-57. doi: 10.1038/nrclinonc.2015.38. Epub 2015 Mar 17. Nat Rev Clin Oncol. 2015. PMID: 25781572 Review.
References
- Hansson EM, Lendahl U, Chapman G. Semin Cancer Biol. 2004;14:320–328. - PubMed
- Allman D, Aster JC, Pear WS. Immunol Rev. 2002;187:75–86. - PubMed
- Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. Cell. 1991;66:649–661. - PubMed
- Weng AP, Ferrando AA, Lee W, Morris JP IV, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Science. 2004;306:269–271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HG002668-03/HG/NHGRI NIH HHS/United States
- T15 LM007079/LM/NLM NIH HHS/United States
- 5 T15 LM007079-13/LM/NLM NIH HHS/United States
- 1U54CA121852-01A1/CA/NCI NIH HHS/United States
- U54 CA121852/CA/NCI NIH HHS/United States
- R01 AI066116/AI/NIAID NIH HHS/United States
- R01 HG002668/HG/NHGRI NIH HHS/United States
- 1R01CA109755-01/CA/NCI NIH HHS/United States
- R01 CA109755/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical