The utility of ETD mass spectrometry in proteomic analysis - PubMed (original) (raw)
Review
The utility of ETD mass spectrometry in proteomic analysis
Leann M Mikesh et al. Biochim Biophys Acta. 2006 Dec.
Abstract
Mass spectrometry has played an integral role in the identification of proteins and their post-translational modifications (PTM). However, analysis of some PTMs, such as phosphorylation, sulfonation, and glycosylation, is difficult with collision-activated dissociation (CAD) since the modification is labile and preferentially lost over peptide backbone fragmentation, resulting in little to no peptide sequence information. The presence of multiple basic residues also makes peptides exceptionally difficult to sequence by conventional CAD mass spectrometry. Here we review the utility of electron transfer dissociation (ETD) mass spectrometry for sequence analysis of post-translationally modified and/or highly basic peptides. Phosphorylated, sulfonated, glycosylated, nitrosylated, disulfide bonded, methylated, acetylated, and highly basic peptides have been analyzed by CAD and ETD mass spectrometry. CAD fragmentation typically produced spectra showing limited peptide backbone fragmentation. However, when these peptides were fragmented using ETD, peptide backbone fragmentation produced a complete or almost complete series of ions and thus extensive peptide sequence information. In addition, labile PTMs remained intact. These examples illustrate the utility of ETD as an advantageous tool in proteomic research by readily identifying peptides resistant to analysis by CAD. A further benefit is the ability to analyze larger, non-tryptic peptides, allowing for the detection of multiple PTMs within the context of one another.
Figures
Figure 1
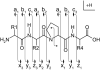
Roepstorff Nomenclature Scheme Illustration of fragment ions formed from the backbone cleavage of protonated peptides. Fragment ions retaining the positive charge on the amino terminus are termed a-, b-, or c-type ions. Fragment ions retaining the positive charge on the carboxy terminus are termed x-, y-, or z-type ions.
Figure 2
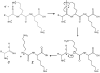
ETD fragmentation scheme Fragmentation scheme of a multiply protonated peptide after reaction with a low energy electron to produce c- and z-type ions [12] Copyright (2004) National Academy of Sciences, USA. Reprinted with permission.
Figure 3
Comparison of CAD vs. ETD spectrum of a phosphorylated peptide Consecutive single-scan CAD vs. ETD mass spectrum comparison of phosphorylated peptides generated from a tryptic digest of human nuclear proteins recorded during a data-dependent analysis (nHPLC-μESI-MS/MS). All peptides were converted to methyl esters and enriched for phosphorylated peptides by immobilized metal affinity chromatography before analysis. (A) CAD spectrum dominated by fragment ions corresponding to the loss of phosphoric acid and either methanol or water. (B) ETD spectrum containing a near complete series of c- and z-type product ions. Note that the spectrum is devoid of fragment ions corresponding to the loss of phosphoric acid [12] Copyright (2004) National Academy of Sciences, USA. Reprinted with permission.
Figure 4
Comparison of CAD vs. ETD spectrum of a sulfonated peptide Sulfonation of peptides was achieved by reacting the peptide with 5% chlorosulfonic acid in neat trifluoroacetic acid (TFA) for 20 minutes at room temperature. The reaction was terminated by the addition of water and was purified by RP-HPLC. Mass spectrometry analysis before and after sulfonation confirmed reaction. Sulfonated peptides (1pmol/ul) were infused at a flow rate of 60 nl/min into a ThermoElectron LTQ ion trap mass spectrometer modified to perform ETD. (A) The CAD spectrum of the illustrated sulfonated peptide contains one major ion corresponding to the neutral loss of SO3 from the (M+3H)+3 precursor ion. (B) Magnification of the spectrum shown in A by 50X. (C) Fragmentation of the sulfonated peptide by ETD where a complete c and z-type ion series was observed with no detectable loss of SO3 from the precursor ion or the peptide backbone.
Figure 5
Comparison of CAD vs. ETD spectrum of O-GlcNAc containing peptide O-GlcNAc containing peptides (1pmol/ul) donated by Gerry Hart were infused at a flow rate of 60 nl/min into a ThermoElectron LTQ ion trap mass spectrometer modified to perform ETD. (A) The CAD spectrum of a synthetic O-GlcNAc modified peptide illustrates the loss of an O-GlcNAc oxoniom ion at m/z 204 and the corresponding charge reduced (M+3H)+3 product ion with a loss of 203 (GlcNAc). (B) The ETD spectrum of the O-GlcNAc modified peptide illustrates an almost complete c and z-type ion series. The only c and z-type ions not observed corresponds to bonds adjacent to a proline residue. In the ETD spectrum of this synthetic peptide, there is no observable loss of GlcNAc.
Figure 6
Comparison of CAD vs. ETD spectrum of an N-linked glycosylated peptide Spectrum of the N-linked glycopeptide generated from a tryptic digest of the Erythina cristagalli (coral tree) lectin. The average mass of the corresponding glycopeptide is 3002 Da with the following N-linked glycan structure Manα3(Manα6)(Xylβ2)Manβ4GlcNAcβ4(Fucα3)GlcNAc. (A) CAD spectrum of the (M+3H)+3 glycopeptide ion. Note fragmentation is of the glycan structure. (B) ETD spectrum of the (M+3H)+3 glycopeptide. Sulfur dioxide was used as the anion. Note that most fragmentation occurs along the peptide backbone [37] Copyright (2005) American Chemical Society. Reprinted with permission.
Figure 7
Comparison of CAD vs. ETD spectrum of a nitrosylated peptide The beta chain of insulin containing two cysteine residues was S-nitrosylated using S-Nitroso-N-acetylpenicillamine (SNAP). The resulting nitrosylated polypeptide was analyzed by nanoflow RP-HPLC interfaced to a ThermoElectron LTQ ion trap mass spectrometer modified to perform ETD. (A) CAD spectrum of the (M+5H)+5 nitrosylated insulin beta chain is mostly composed of the neutral loss of both NO groups on the cysteine residues of insulin. Minimal peptide backbone fragmentation is observed. (B) ETD spectrum of nitrosylated insulin beta chain illustrates a charge reduced (electron transfer without fragmentation) species with and without losses of NO. However, a few low level c and z-type ions demonstrate the retention of NO on some product ions after ETD.
Figure 8
Comparison of CAD vs. ETD spectrum of histone H3 peptides Human histone H3 peptides (amino acids 6-20 or 6-19) were synthesized on an AMS 422 multiple peptide synthesizer (Gilson Medical Electronics, Middletown, WI) by standard solid-phase 9-fluorenylmethoxycarbonyl (Fmoc) chemistry using Wang resins. Novabiochem Fmoc protected amino acids, modified amino acids (phosphorylated, acetylated, and methylated), and resins were used for peptide synthesis. Synthetic histone peptides (1pmol/ul) were infused at a flow rate of 60 nl/min into a ThermoElectron LTQ ion trap mass spectrometer modified to perform ETD. (A) The CAD spectrum from histone H3 (6-20), TAR(acK)STGGKAPRKQL, is illustrated. The spectrum consists of a product ion corresponding to the neutral loss of water with minimal peptide backbone fragmentation. (B) The corresponding ETD spectrum illustrates an almost complete c and z-type ion series for the peptide. The only fragment ions not observed are those N-terminal to proline (due to the ring structure of proline), z-type ions where there is no amino acid residue to be protonated at the C-terminus (z1), and c1.
Figure 9
Tandem mass spectrum of ubiquitin generated by sequential ion/ion reactions (A) Spectrum illustrating whole protein dissociation of ubiquitin, a 8.5 kDa protein (+13, m/z 659), after a 15-ms reaction with the radical anion of fluoranthene. Several hundred highly charged unresolved c- and z-type fragment ions dominate the spectrum. (_B_-D) Sequestering the entire mixture of highly charged product ions and reacting them with a second anion, deprotonated benzoic acid, simplified this mixture. Note the gradual degradation of multiply charged products when reacted with even-electron anions of benzoic acid for 50 (B), 100 (C), and 150 (D) ms., leaving predominately doubly and singly charged fragments after 150 ms. (E) The resulting sequence coverage considering only singly charged product ions. Each spectrum is the average of ∼50 spectra (∼30-s acquisition), and the y axis indicates the relative ion abundance [30] Copyright (2005) National Academy of Sciences, USA. Reprinted with permission.
Figure 10
Sequencing highly modified, large peptides by ETD Online chromatographic separation of large peptides (residues 1-50 from histone H3.1) followed by automated sequential ion/ion reactions of 15-ms ETD followed by 150-ms PTR. (A) The tandem mass spectrum from early eluting histone H3.1 peptide. Note the c- and z-type ion series allows interpretation of the N- and C-termini and demonstrates that K and K 4 9 are modified with monomethyl and dimethyl groups respectively. (B) The tandem mass spectrum generated from a histone H3.1 peptide eluting 6 seconds later. Here the c-type ion series indicates the N terminus is modified identical to the peptide shown in (A); however, the C terminus is monomethylated at K37, as opposed to being dimethylated at K36. Note the spectrum shown in (B) contains fragment ions derived from both species (co-elution) [30] Copyright (2005) National Academy of Sciences, USA. Reprinted with permission.
Similar articles
- Quantification of post-translationally modified peptides of bovine alpha-crystallin using tandem mass tags and electron transfer dissociation.
Viner RI, Zhang T, Second T, Zabrouskov V. Viner RI, et al. J Proteomics. 2009 Jul 21;72(5):874-85. doi: 10.1016/j.jprot.2009.02.005. Epub 2009 Feb 24. J Proteomics. 2009. PMID: 19245863 - Application of electron transfer dissociation (ETD) for the analysis of posttranslational modifications.
Wiesner J, Premsler T, Sickmann A. Wiesner J, et al. Proteomics. 2008 Nov;8(21):4466-83. doi: 10.1002/pmic.200800329. Proteomics. 2008. PMID: 18972526 Review. - Recent developments and applications of electron transfer dissociation mass spectrometry in proteomics.
Sarbu M, Ghiulai RM, Zamfir AD. Sarbu M, et al. Amino Acids. 2014 Jul;46(7):1625-34. doi: 10.1007/s00726-014-1726-y. Epub 2014 Apr 1. Amino Acids. 2014. PMID: 24687149 Review. - Electron capture dissociation mass spectrometry in characterization of peptides and proteins.
Bakhtiar R, Guan Z. Bakhtiar R, et al. Biotechnol Lett. 2006 Jul;28(14):1047-59. doi: 10.1007/s10529-006-9065-z. Epub 2006 Jun 23. Biotechnol Lett. 2006. PMID: 16794768 Review.
Cited by
- Defining the disulfide bonds of insulin-like growth factor-binding protein-5 by tandem mass spectrometry with electron transfer dissociation and collision-induced dissociation.
Nili M, Mukherjee A, Shinde U, David L, Rotwein P. Nili M, et al. J Biol Chem. 2012 Jan 6;287(2):1510-9. doi: 10.1074/jbc.M111.285528. Epub 2011 Nov 22. J Biol Chem. 2012. PMID: 22117064 Free PMC article. - Gas-Phase Oxidation of Neutral Basic Residues in Polypeptide Cations by Periodate.
Pilo AL, Bu J, McLuckey SA. Pilo AL, et al. J Am Soc Mass Spectrom. 2016 Dec;27(12):1979-1988. doi: 10.1007/s13361-016-1491-0. Epub 2016 Sep 19. J Am Soc Mass Spectrom. 2016. PMID: 27644939 Free PMC article. - Leveraging Immonium Ions for Targeting Acyl-Lysine Modifications in Proteomic Datasets.
Muroski JM, Fu JY, Nguyen HH, Ogorzalek Loo RR, Loo JA. Muroski JM, et al. Proteomics. 2021 Feb;21(3-4):e2000111. doi: 10.1002/pmic.202000111. Epub 2020 Sep 25. Proteomics. 2021. PMID: 32896103 Free PMC article. - Bottom-up and middle-down proteomics have comparable accuracies in defining histone post-translational modification relative abundance and stoichiometry.
Sidoli S, Lin S, Karch KR, Garcia BA. Sidoli S, et al. Anal Chem. 2015 Mar 17;87(6):3129-33. doi: 10.1021/acs.analchem.5b00072. Epub 2015 Mar 3. Anal Chem. 2015. PMID: 25719549 Free PMC article. - Modulation of 14-3-3 interaction with phosphorylated histone H3 by combinatorial modification patterns.
Winter S, Fischle W, Seiser C. Winter S, et al. Cell Cycle. 2008 May 15;7(10):1336-42. doi: 10.4161/cc.7.10.5946. Epub 2008 Mar 11. Cell Cycle. 2008. PMID: 18418070 Free PMC article.
References
- Crane R, Gadea B, Littlepage L, Wu H, Ruderman JV. Aurora A, Meiosis and Mitosis. Biology of the Cell. 2004;96:215–229. - PubMed
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. - PubMed
- Li N, Lerea KM, Etlinger JD. Phosphorylation of the Proteasome Activator PA28 Is Required for Proteasome Activation. Biochemical and Biophysical Research Communications. 1996;225:855–860. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous