Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes - PubMed (original) (raw)
Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes
Kevin Kim et al. RNA. 2007 Jan.
Abstract
In the Drosophila RNA interference (RNAi) pathway, small interfering RNAs (siRNAs) direct Argonaute2 (Ago2), an endonuclease, within the RNA-induced silencing complex (RISC) to cleave complementary mRNA targets. In vitro studies have shown that, for each siRNA duplex, RISC retains only one strand, the guide, and releases the other, the passenger, to form a holo-RISC complex. Here, we have isolated a new Ago2 mutant allele and provide, for the first time, in vivo evidence that endogenous Ago2 slicer activity is important to mount an RNAi response in Drosophila. We demonstrate in vivo that efficient removal of the passenger strand from RISC requires the cleavage activity of Ago2. We have also identified a new intermediate complex in the RISC assembly pathway, pre-RISC, in which Ago2 is stably bound to double-stranded siRNA.
Figures
FIGURE 1.
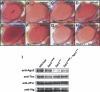
Phenotype of the Ago2V966M MutantCompound eyes from white+ adult flies (A–H). (A) An Ago2+ adult. (B) An Ago2+ adult with one copy of GMR-wIR. The white gene is partially silenced, resulting in a pale orange eye color. (C) An adult with one copy of GMR-wIR and heterozygous for Ago2V966M. (D) An adult with two copies of GMR-wIR and homozygous for Ago2V966M. (E) An adult with two copies of GMR-wIR and homozygous for Ago2414. (F) An adult with two copies of GMR-wIR and _trans_-heterozygous for Ago2V966M and Ago2414. (G) An Ago2414 adult homozygote with two copies of GMR-wIR and a single copy of the Ago2 rescue transgene. (H) An Ago2V966M adult homozygote with two copies of GMR-wIR and a single copy of the Ago2 rescue transgene. (I) Lysates prepared from wild-type, Ago2V966M, Ago2414, and Ago2V966M/Ago2414 embryos were separated by SDS-PAGE, blotted onto nitrocellulose membrane, and subjected to Western analysis using antibodies against TSN, VIG, dFXR, and Ago2.
FIGURE 2.
Structural characteristics of Ago2V966M. (A) Protein sequence alignment of Argonaute Piwi domains of Drosophila (Dm), Human (Hs), Pyrococcus furiosus (Pf). Aligned sequences are color-coded: 100% conserved residues (black), 75% conserved residues (light gray), and well-conserved hydrophobic or hydrophilic residues (dark gray). The two conserved motifs, GxDV and RDG, are indicated with a bar below the sequence. Three conserved catalytic residues within the Piwi domain are marked with circles at the top of the alignment. The mutation in Ago2V966M is marked with an arrow above the alignment. (B) Schematic representation of Drosophila Ago2 protein with known domains. The mutation of Ago2V966M is labeled.
FIGURE 3.
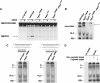
Biochemical characterization of Ago2V966M. (A) In vitro target RNA cleavage assay. Lysates from wild-type, Ago2V966M, Ago2414, and Ago2V966M/Ago2414 embryos were incubated with 5′-labeled _Pp_-luc target mRNA and unlabeled cognate siRNA duplex, as indicated. The 5′ product from RISC activity is indicated and comigrates on the gel with a cleavage product from siRNA-independent nuclease activity. (B) RISC formation assay. Lysates from wild-type and Ago2 mutant embryos were incubated with labeled _Pp_-luc siRNA, and complexes were resolved by native gel electrophoresis. RDI and RLC intermediate complexes are indicated. The complex (arrow) closely corresponding to holo-RISC in Ago2V966M mutant was of slower electrophoretic mobility when compared with wild-type holo-RISC. Ago2V966M/Ago2414 embryo lysate showed reduced abundance of the slower complex. (C) RISC formation with a highly asymmetric siRNA. Either the guide (top) or the passenger (bottom) strand was 5′-radiolabeled. Both wild-type and Ago2V966M lysates form two discrete low-mobility complexes when incubated with radiolabeled guide strand. In wild-type lysate, the slower complex (S) was less abundant than the faster complex (F). In Ago2V966M lysate, the relative amount of S complex increased while F complex decreased. When incubated with radiolabeled passenger strand, both wild-type and Ago2V966M lysates formed only the S complex due to asymmetric loading of unlabeled guide strand into holo-RISC. Note that more of the S complex was formed in Ago2V966M lysate. (D) Radiolabeled guide siRNA strand was incubated with either unlabeled cognate or noncognate 2′-_O_-methyl target mRNA in wild-type lysate. Addition of cognate target mRNA resulted in the depletion of the F complex, whereas the S complex was not affected. Addition of noncognate mRNA had no effect on either complex.
FIGURE 4.
Ago2V966M RISC complexes contain primarily duplex siRNA. (A) Schematic representation of two-dimensional gel electrophoresis. RISC formation reaction was performed with guide-labeled siRNA duplex. The complexes were resolved in the first dimension by native gel electrophoresis. The entire lane was than excised and placed horizontally on a 15% SDS-polyacrylamide gel. Labeled siRNAs from complexes were then subjected to electrophoresis in the second dimension. (B) Both wild-type and Ago2V966M lysates show RDI and RLC complexes that contain duplex siRNA. The low-mobility complexes observed in wild-type lysate primarily contain single-stranded siRNA while those formed in Ago2V966M lysate primarily contain duplex siRNA.
FIGURE 5.
Ago2V966M is defective in siRNA duplex dissociation. (A) Guide-labeled siRNA duplex was incubated in wild-type, Ago2V966M, or Ago2414 lysate. Reactions were resolved on a 15% SDS-polyacrylamide gel and radioactive signal was quantified by PhosphorImager. The percentage of single-stranded siRNA generated from siRNA duplex was plotted against reaction time. (B) Passenger-labeled siRNA duplex was incubated in the same lysates as A. Reactions at various time points were removed, deproteinized, and electrophoresed to visualize passenger strand cleavage products. The 9-nt 5′ cleavage product accumulated early in the reaction and disappeared. Ago2V966M mutant lysate produced little passenger strand cleavage product. The percentage of the 9-nt product cleaved from labeled passenger strand was plotted against reaction time.
FIGURE 6.
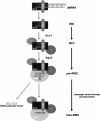
Model for RISC assembly in Drosophila in vitro. The siRNA is bound to R2D2/Dcr2 heterodimer to form a RDI complex. The RDI complex then enters a RISC-loading complex (RLC), associating with other RNAi components. RLC then recruits Ago2 to form a pre-RISC complex that contains duplex siRNA. Endonucleolytic activity of Ago2 within the Piwi domain promotes release of the passenger strand from pre-RISC to form holo-RISC. Holo-RISC can then base-pair to complementary single-stranded mRNA substrates for cleavage.
Similar articles
- Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes.
Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Matranga C, et al. Cell. 2005 Nov 18;123(4):607-20. doi: 10.1016/j.cell.2005.08.044. Epub 2005 Nov 3. Cell. 2005. PMID: 16271386 - Slicer function of Drosophila Argonautes and its involvement in RISC formation.
Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Miyoshi K, et al. Genes Dev. 2005 Dec 1;19(23):2837-48. doi: 10.1101/gad.1370605. Epub 2005 Nov 14. Genes Dev. 2005. PMID: 16287716 Free PMC article. - [Components and assembly of RNA-induced silencing complex].
Song XM, Yan F, Du LX. Song XM, et al. Yi Chuan. 2006 Jun;28(6):761-6. Yi Chuan. 2006. PMID: 16818443 Review. Chinese. - Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways.
Okamura K, Ishizuka A, Siomi H, Siomi MC. Okamura K, et al. Genes Dev. 2004 Jul 15;18(14):1655-66. doi: 10.1101/gad.1210204. Epub 2004 Jul 1. Genes Dev. 2004. PMID: 15231716 Free PMC article. - The RNAi pathway initiated by Dicer-2 in Drosophila.
Kim K, Lee YS, Harris D, Nakahara K, Carthew RW. Kim K, et al. Cold Spring Harb Symp Quant Biol. 2006;71:39-44. doi: 10.1101/sqb.2006.71.008. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381278 Review.
Cited by
- Homology directed repair is unaffected by the absence of siRNAs in Drosophila melanogaster.
Schmidts I, Böttcher R, Mirkovic-Hösle M, Förstemann K. Schmidts I, et al. Nucleic Acids Res. 2016 Sep 30;44(17):8261-71. doi: 10.1093/nar/gkw570. Epub 2016 Jun 27. Nucleic Acids Res. 2016. PMID: 27353331 Free PMC article. - Two new and distinct roles for Drosophila Argonaute-2 in the nucleus: alternative pre-mRNA splicing and transcriptional repression.
Taliaferro JM, Aspden JL, Bradley T, Marwha D, Blanchette M, Rio DC. Taliaferro JM, et al. Genes Dev. 2013 Feb 15;27(4):378-89. doi: 10.1101/gad.210708.112. Epub 2013 Feb 7. Genes Dev. 2013. PMID: 23392611 Free PMC article. - QIP, a putative exonuclease, interacts with the Neurospora Argonaute protein and facilitates conversion of duplex siRNA into single strands.
Maiti M, Lee HC, Liu Y. Maiti M, et al. Genes Dev. 2007 Mar 1;21(5):590-600. doi: 10.1101/gad.1497607. Epub 2007 Feb 20. Genes Dev. 2007. PMID: 17311884 Free PMC article. - Argonaute2 suppresses Drosophila fragile X expression preventing neurogenesis and oogenesis defects.
Pepper AS, Beerman RW, Bhogal B, Jongens TA. Pepper AS, et al. PLoS One. 2009 Oct 27;4(10):e7618. doi: 10.1371/journal.pone.0007618. PLoS One. 2009. PMID: 19888420 Free PMC article. - Proteomics identification of Drosophila small interfering RNA-associated factors.
Gerbasi VR, Golden DE, Hurtado SB, Sontheimer EJ. Gerbasi VR, et al. Mol Cell Proteomics. 2010 Sep;9(9):1866-72. doi: 10.1074/mcp.M900614-MCP200. Epub 2010 May 15. Mol Cell Proteomics. 2010. PMID: 20472918 Free PMC article.
References
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. - PubMed
- Lee, Y.S., Carthew, R.W. Making a better RNAi vector for Drosophila: Use of intron spacers. Methods. 2003;30:322–329. - PubMed
- Lee, Y.S., Nakahara, K., Pham, J.W., Kim, K., He, Z., Sontheimer, E.J., Carthew, R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. - PubMed
- Liu, Q., Rand, T.A., Kalidas, S., Du, F., Kim, H.E., Smith, D.P., Wang, X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases