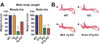Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice - PubMed (original) (raw)
Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice
Erin L McDearmon et al. Science. 2006.
Abstract
The basic helix-loop-helix (bHLH)-Per-Arnt-Sim (PAS) domain transcription factor BMAL1 is an essential component of the mammalian circadian pacemaker. Bmal1-/- mice lose circadian rhythmicity but also display tendon calcification and decreased activity, body weight, and longevity. To investigate whether these diverse functions of BMAL1 are tissue-specific, we produced transgenic mice that constitutively express Bmal1 in brain or muscle and examined the effects of rescued gene expression in Bmal1-/- mice. Circadian rhythms of wheel-running activity were restored in brain-rescued Bmal1-/- mice in a conditional manner; however, activity levels and body weight were lower than those of wild-type mice. In contrast, muscle-rescued Bmal1-/- mice exhibited normal activity levels and body weight yet remained behaviorally arrhythmic. Thus, Bmal1 has distinct tissue-specific functions that regulate integrative physiology.
Figures
Fig. 1
_Bmal1_-containing BAC transgenes rescue _Bmal1_−/− phenotypes. (A) Bmal1 BAC clones were used to create transgenic (Tg) mice, which were consecutively crossed with Bmal1+/− mice to create BAC-rescued _Bmal1_−/− mice. (B) Bmal1 mRNA levels in SCN were examined by in situ hybridization in wild-type (WT) and Bmal1 BAC Tg mice, sacrificed at ZT6 and ZT18 (shown are mean ± SEM; significant effect of genotype, GLM ANOVA). (C) Representative wheel-running activity records from WT, Bmal1 BAC Tg, _Bmal1_−/− (Bmal1 KO) or BAC-rescued (Bmal1 BAC Tg; Bmal1 KO) mice. Mice were housed in LD then released into DD for 3 weeks. (D) Bar graphs of mean ± SEM show that BAC-rescued mice (n=6) exhibit free-running period, amplitude of circadian rhythm and activity levels that are not significantly different from WT. Amplitude is graphed as the peak amplitude of the proportion of the total variance in the time series in the circadian (~24h) range (***p<0.001, one-way ANOVA; *significant effect of genotype, GLM ANOVA).
Fig. 2
Reversible restoration of circadian rhythms but not activity levels in brain-rescued _Bmal1_−/− mice. (A) Mice were created to express Bmal1-HA conditionally in brain tissue by using the tTA system. (B) In situ hybridization was performed with HA tag or Bmal1 probes on brains from WT, tetO∷Bmal1-HA (tetO), or Scg2∷tTA x tetO∷Bmal1-HA double transgenic (DTg) mice sacrificed at ZT 6 (arrow indicates SCN, scale bar is 1mm). (C) Western blot showing HA staining in brain, liver and skeletal muscle protein extracts from WT or DTg mice sacrificed at ZT 12 (arrow indicates correct size of BMAL1). (D) Representative wheel-running activity records from one _Bmal1_−/− mouse and two brain-rescued _Bmal1_−/− (DTg; Bmal1 KO) mice. After 3 weeks in DD (Pre), mice were administered Dox for 2 weeks (Dox, highlighted yellow) and then spent an additional 3 weeks without Dox (Post). (E) Brain-rescued mice (n=10) display a free-running period of 22.8 h (Pre) and 22.6 h (Post) when Bmal1 is expressed, which are significantly different from WT and DTg groups (*significant effect of genotype, GLM ANOVA). Activity levels of KO and DTg KO mice were significantly reduced compared to WT. Amplitude of circadian rhythm was significantly different in all genotypes compared to WT, and a simultaneous loss of rhythm and decrease in amplitude were observed in DTg KO mice during Dox treatment (†significant effect of time interval). Graphs represent the mean ± SEM.
Fig. 3
Muscle-rescued mice exhibit restored activity level but not circadian rhythms. (A) Muscle-specific Bmal1 Tg mice were created by fusing the Acta1 promoter sequence to Bmal1-HA. (B) Western blot shows HA staining in brain, liver and skeletal muscle protein extracts from WT or Tg mice sacrificed at ZT 12 (arrow indicates correct size of BMAL1). (C) Representative wheel-running activity records are shown from WT, Tg, Bmal1 KO, and muscle-rescued (Tg; Bmal1 KO) mice. (D) Muscle-rescued mice (n=6) are arrhythmic in DD with significantly reduced amplitude of rhythm (**p<0.01, one-way ANOVA), but display activity levels that are not significantly different from WT mice. Graphs show mean ± SEM (*significant effect of genotype, GLM ANOVA).
Fig. 4
Effects of tissue-specific Bmal1 expression on body weight and tendon calcification. (A) Brain-rescued mice and KO mice in both lines have significantly reduced body weight, while muscle-rescued mice exhibit similar body weight to WT mice (graphs represent mean + SEM; **p<0.01 ***p<0.001, one-way ANOVA). (B) Photographs of Alizarin Red-stained hind limbs from WT, KO, muscle and brain-rescued KO mice are shown. Arrows indicate calcaneal tendon calcification in all but WT mice.
Similar articles
- Differential rescue of light- and food-entrainable circadian rhythms.
Fuller PM, Lu J, Saper CB. Fuller PM, et al. Science. 2008 May 23;320(5879):1074-7. doi: 10.1126/science.1153277. Science. 2008. PMID: 18497298 Free PMC article. - New reporter system for Per1 and Bmal1 expressions revealed self-sustained circadian rhythms in peripheral tissues.
Nishide SY, Honma S, Nakajima Y, Ikeda M, Baba K, Ohmiya Y, Honma K. Nishide SY, et al. Genes Cells. 2006 Oct;11(10):1173-82. doi: 10.1111/j.1365-2443.2006.01015.x. Genes Cells. 2006. PMID: 16999737 - Suprachiasmatic regulation of circadian rhythms of gene expression in hamster peripheral organs: effects of transplanting the pacemaker.
Guo H, Brewer JM, Lehman MN, Bittman EL. Guo H, et al. J Neurosci. 2006 Jun 14;26(24):6406-12. doi: 10.1523/JNEUROSCI.4676-05.2006. J Neurosci. 2006. PMID: 16775127 Free PMC article. - [Synchronization and genetic redundancy in circadian clocks].
Dardente H. Dardente H. Med Sci (Paris). 2008 Mar;24(3):270-6. doi: 10.1051/medsci/2008243270. Med Sci (Paris). 2008. PMID: 18334175 Review. French. - [BMAL1 and circadian rhythm].
Ikeda M. Ikeda M. Nihon Shinkei Seishin Yakurigaku Zasshi. 2000 Nov;20(5):203-12. Nihon Shinkei Seishin Yakurigaku Zasshi. 2000. PMID: 11326546 Review. Japanese.
Cited by
- Neuromedin s-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms.
Lee IT, Chang AS, Manandhar M, Shan Y, Fan J, Izumo M, Ikeda Y, Motoike T, Dixon S, Seinfeld JE, Takahashi JS, Yanagisawa M. Lee IT, et al. Neuron. 2015 Mar 4;85(5):1086-102. doi: 10.1016/j.neuron.2015.02.006. Neuron. 2015. PMID: 25741729 Free PMC article. - Cell state dependent effects of Bmal1 on melanoma immunity and tumorigenicity.
Zhang X, Pant SM, Ritch CC, Tang HY, Shao H, Dweep H, Gong YY, Brooks R, Brafford P, Wolpaw AJ, Lee Y, Weeraratna A, Sehgal A, Herlyn M, Kossenkov A, Speicher D, Sorger PK, Santagata S, Dang CV. Zhang X, et al. Nat Commun. 2024 Jan 20;15(1):633. doi: 10.1038/s41467-024-44778-2. Nat Commun. 2024. PMID: 38245503 Free PMC article. - Stable isotope ratios in hair and teeth reflect biologic rhythms.
Appenzeller O, Qualls C, Barbic F, Furlan R, Porta A. Appenzeller O, et al. PLoS One. 2007 Jul 25;2(7):e636. doi: 10.1371/journal.pone.0000636. PLoS One. 2007. PMID: 17653263 Free PMC article. - Translational switching of Cry1 protein expression confers reversible control of circadian behavior in arrhythmic Cry-deficient mice.
Maywood ES, Elliott TS, Patton AP, Krogager TP, Chesham JE, Ernst RJ, Beránek V, Brancaccio M, Chin JW, Hastings MH. Maywood ES, et al. Proc Natl Acad Sci U S A. 2018 Dec 26;115(52):E12388-E12397. doi: 10.1073/pnas.1811438115. Epub 2018 Nov 28. Proc Natl Acad Sci U S A. 2018. PMID: 30487216 Free PMC article. - Disruption of the mouse Bmal1 locus promotes heterotopic ossification with aging via TGF-beta/BMP signaling.
Liang Q, Lu Y, Yu L, Zhu Q, Xie W, Wang Y, Ye L, Li Q, Liu S, Liu Y, Zhu C. Liang Q, et al. J Bone Miner Metab. 2022 Jan;40(1):40-55. doi: 10.1007/s00774-021-01271-w. Epub 2021 Oct 9. J Bone Miner Metab. 2022. PMID: 34626248
References
- Ikeda M, Nomura M. Biochem Biophys Res Commun. 1997;233:258. - PubMed
- Hogenesch JB, et al. J Biol Chem. 1997;272:8581. - PubMed
- Gekakis N, et al. Science. 1998;280:1564. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases