Combinatorial function of ETS transcription factors in the developing vasculature - PubMed (original) (raw)
Combinatorial function of ETS transcription factors in the developing vasculature
Van N Pham et al. Dev Biol. 2007.
Abstract
Members of the ETS family of transcription factors are among the first genes expressed in the developing vasculature, but loss-of-function experiments for individual ETS factors in mice have not uncovered important early functional roles for these genes. However, multiple ETS factors are expressed in spatially and temporally overlapping patterns in the developing vasculature, suggesting possible functional overlap. We have taken a comprehensive approach to exploring the function of these factors during vascular development by employing the genetic and experimental tools available in the zebrafish to analyze four ETS family members expressed together in the zebrafish vasculature; fli1, fli1b, ets1, and etsrp. We isolated and characterized an ENU-induced mutant with defects in trunk angiogenesis and positionally cloned the defective gene from this mutant, etsrp. Using the etsrp morpholinos targeting each of the four genes, we show that the four ETS factors function combinatorially during vascular and hematopoietic development. Reduction of etsrp or any of the other genes alone results in either partial or no defects in endothelial differentiation, while combined reduction in the function of all four genes causes dramatic loss of endothelial cells. Our results demonstrate that combinatorial ETS factor function is essential for early endothelial specification and differentiation.
Figures
Figure 1
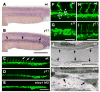
y11 mutants have defects in angiogenesis and vascular morphogenesis. (A,B) TUNEL staining of the trunks of 24 hpf wild type (A) and y11 mutant (B) animals, low background in wild type animals and substantially increased staining in the developing axial vessels of y11 mutants (arrows). (C–I) Two-photon images of Tg(fli1:EGFP)y1 transgenic wild type or y11 mutant animals at 24 hpf (C–G) and 48 hpf (H,I). Wild type siblings (C,F,H) have intersegmental vessel sprouts (arrows in C) and properly lumenized axial vessels (arrowheads in F) at 24 hpf. Intersegmental vessels have formed a lumenized, functional vascular network by 48 hpf (H). In y11 mutants (D,G,I) intersegmental vessel sprouts are absent at 24 hpf (D) and axial vessels fail to undergo proper tubular morphogenesis (G). Intersegmental vessels are similarly absent in etsrp morphants at 24 hpf (E). By 48 hpf y11 mutants have aberrant intersegmental vessel sprouts that are not fully extended and have some branching and pathfinding errors (I). (J,K) Electron microscopy of 24 hpf zebrafish shows defects in vascular morphogenesis in y11 mutants. Wild type siblings have normal single-cell thick endothelial epithelium (arrows) around the dorsal aorta (J), but endothelial cells in y11 mutants do not form a proper epithelium (K). Anterior is to the left, dorsal up in all panels. Rostral is to the left and dorsal is up in panels A–I. Scale bars = 200 μm (A,B), 200 μm (C–E), 50 μm (F,G), 100 μm (H,I), 10 μm (J,K)
Figure 2
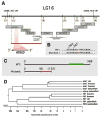
Positional cloning of etsrp, the defective gene in y11 mutants. (A) Genetic and physical map of the y11 interval. Genetic map is shown at top, with the number of recombinants in approximately 2000 meioses noted in brown (and whether recombinants are on the right or left sides, in parentheses). BAC and PAC clones are shown as large grey bars, genomic sequence contigs and traces are shown as black bars. The red bar shows the y11 critical interval containing the entire etsrp transcript. (B) In y11 mutants a G is inserted after nucleotide 269 in the etsrp coding sequence. (C) The y11 insertion mutation results in an altered reading frame after amino acid 90 of etsrp coding for an additional novel 62 amino acids (shown in red) before terminating at a stop codon. The y11 mutant protein lacks the highly conserved ETS DNA binding domain (shown in green in the wild type protein). See results and methods for additional details. (D) ETS family members expressed in the zebrafish vasculature and related mammalian orthologs. Dendogram of nucleotide simiilarity across the entire coding region of zebrafish ETS family members expressed in the zebrafish vasculature and the most closely related mammalian genes. The zebrafish ets1 and fli1 genes are more closely related to their mammalian counterparts than to other zebrafish ETS family members. The etsrp and fli1b genes are most closely related to ets1 and fli1 genes, respectively.
Figure 3
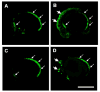
Induction of ectopic fli1-EGFP and flk1-GFP expression by etsrp mRNA injection. (A–D) Confocal images of Tg(fli1:EGFP)y1 (A,B) and Tg(flk1:GFP) (C,D) transgenic animals at approximately 14 somite stage/16 hours post-fertilization. (A,C) Uninjected control animals. (B,D) Animals injected at the 4–8 cell stage with 20 pg etsrp mRNA, targeting a limited number of blastomeres. Ectopic expression of the fli1-EGFP and flk1-GFP transgenes is noted by large arrows; small arrows show normal expression domains. Rostral is to the left and dorsal is up in all panels. Scale bar = 400 μm.
Figure 4
Expression of zebrafish ETS transcription factors during somitogenesis, with flk1 and vecdn1 shown for comparison. Whole mount in situ hybridization using probes for zebrafish etsrp (A, E, K, Q, W), ets1 (B, F, L, R, X), fli1 (C, G, M, S, Y), fli1b (D, H, N, T, Z), flk1 (I, O, U, A’), and ve-cdn (J, P, V, B’). Embryos are probed at the 5 somite stage (ss)/11.5 hours post-fertilization (hpf) (A–D), 10 ss/14 hpf (E–J), 15 ss/16.5 hpf (K–P), and 24 hours post-fertilization (Q-B’). Images in A–P are complete rostral to caudal collages of multiple dorsal-view images, for a “virtual flat mount.” Images in Q–V are lateral views of the entire 24 hpf animal, while W-B’ are higher magnification lateral views of 24 hpf trunks. Rostral is to the left in all panels, dorsal is up in Q-B’. Scale bars = 1000 μm (A–V), and 250 μm (W–B’).
Figure 5
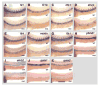
Expression of vascular marker genes in y11 mutants at 14 hpf (approximately 10 somite stage). Panels show whole mount in situ hybridization of 14 hpf wild type siblings (A–D) and y11 mutants (E–H) probed for flk1 (A,E), flt4 (B,F,I), vecdn (C,G), and plxnd1 (D,H). The developing trunk region imaged in panels A–H is noted by the box on the whole _flt4_-stained wild type embryo in panel I. Rostral is up in all panels. Scale bar = 200 μm (A–H).
Figure 6
Expression of ETS transcription factors and vascular and hematopoietic marker genes in y11 mutants at 24 hpf (approximately 30 somite stage). Panels show whole mount in situ hybridization of the trunks of 24 hpf wild type siblings (top of each panel) and y11 mutants (bottom of each panel) probed for fli1 (A), fli1b (B), ets1 (C), etsrp (D), flt4 (E), vecdn (F), flk1 (G), plxnd1 (H), efnb2 (I), gata1 (J), and gata2 (K). Rostral is to the left and dorsal is up in all panels. Scale bar = 200 μm.
Figure 7
Trunk circulation and intersegmental vessel sprouting defects in zebrafish injected with morpholinos targeting vascular ETS factors. Morpholinos targeting fli1, fli1b, ets1, or etsrp were injected into Tg(fli1:EGFP)y1 zebrafish either alone, in combinations of three morpholinos, or all together, as noted by “x” marks at the top and bottom of the figure. The number of animals for which phenotypes were assessed for each injection is shown at the extreme bottom of the Fig. (N=). The upper graph shows the % of animals with active blood circulation in the trunk at 24 hpf (purple bars) and 48 hpf (red bars). The middle graph shows the % of animals at 24 hpf with 1–15 (orange bars) or >15 (blue bars) intersegmental vessel sprouts in the trunk proper, as visualized by microscopic examination of fli1:EGFP transgene expression in developing blood vessels. The lower graph shows the % of animals at 48 hpf with 1–15 (orange bars) or >15 (blue bars) intersegmental vessel sprouts in the trunk proper. The graphs also include tabulation of circulation and intersegmental vessel sprout phenotypes measured in control morpholino injected animals, all of which had trunk circulation and a full complement of trunk intersegmental vessel sprouts.
Figure 8
Reduction of all four vascular ETS transcription factors by morpholino knockdown. (A–K) Expression of ETS transcription factors and vascular and hematopoietic marker genes in animals injected with either a control morpholino (top of each panel) or a cocktail of four morpholinos targeting the fli1, fli1b, ets1, and etsrp genes (bottom of each panel). The four morpholino cocktail was injected at the “medium” dose, and an equivalent dosage of morpholino (ng) was injected in control animals. Panels show whole mount in situ hybridization of the trunks of 24 hpf morpholino-injected animals probed for zebrafish fli1 (A), fli1b (B), ets1 (C), etsrp (D), flt4 (E), vecdn (F), flk1 (G), plxnd1 (H), efnb2 (I), gata1 (J), and gata2 (K). (L,M) Confocal images of the mid-trunk of 36 hpf control- (L) and four morpholino cocktail- (M) injected Tg(fli1:nEGFP)y7 animals, collected using identical microscope settings. (L) Control animals display numerous brightly EGFP-positive endothelial nuclei in both axial (arrowheads) and intersegmental (arrows) vessels. (M) Only a small number of residual moderately EGFP-positive cells are seen in the position of the axial vessel (arrows) in four morpholino-injected animals. Anterior is to the left in all images. Scale bar = 250 μm.
Similar articles
- Etv2 and fli1b function together as key regulators of vasculogenesis and angiogenesis.
Craig MP, Grajevskaja V, Liao HK, Balciuniene J, Ekker SC, Park JS, Essner JJ, Balciunas D, Sumanas S. Craig MP, et al. Arterioscler Thromb Vasc Biol. 2015 Apr;35(4):865-76. doi: 10.1161/ATVBAHA.114.304768. Epub 2015 Feb 26. Arterioscler Thromb Vasc Biol. 2015. PMID: 25722433 Free PMC article. - The zebrafish ETS transcription factor Fli1b functions upstream of Scl/Tal1 during embryonic hematopoiesis.
Laverde V, Loges L, Sumanas S. Laverde V, et al. Biol Open. 2025 Apr 15;14(4):bio061948. doi: 10.1242/bio.061948. Epub 2025 Apr 9. Biol Open. 2025. PMID: 40079219 Free PMC article. - Fli+ etsrp+ hemato-vascular progenitor cells proliferate at the lateral plate mesoderm during vasculogenesis in zebrafish.
Chun CZ, Remadevi I, Schupp MO, Samant GV, Pramanik K, Wilkinson GA, Ramchandran R. Chun CZ, et al. PLoS One. 2011 Feb 25;6(2):e14732. doi: 10.1371/journal.pone.0014732. PLoS One. 2011. PMID: 21364913 Free PMC article. - Mouse models in the study of the Ets family of transcription factors.
Bartel FO, Higuchi T, Spyropoulos DD. Bartel FO, et al. Oncogene. 2000 Dec 18;19(55):6443-54. doi: 10.1038/sj.onc.1204038. Oncogene. 2000. PMID: 11175360 Review. - ETS transcription factors in embryonic vascular development.
Craig MP, Sumanas S. Craig MP, et al. Angiogenesis. 2016 Jul;19(3):275-85. doi: 10.1007/s10456-016-9511-z. Epub 2016 Apr 28. Angiogenesis. 2016. PMID: 27126901 Free PMC article. Review.
Cited by
- Etv2 and fli1b function together as key regulators of vasculogenesis and angiogenesis.
Craig MP, Grajevskaja V, Liao HK, Balciuniene J, Ekker SC, Park JS, Essner JJ, Balciunas D, Sumanas S. Craig MP, et al. Arterioscler Thromb Vasc Biol. 2015 Apr;35(4):865-76. doi: 10.1161/ATVBAHA.114.304768. Epub 2015 Feb 26. Arterioscler Thromb Vasc Biol. 2015. PMID: 25722433 Free PMC article. - Zebrafish as a Model for the Study of Human Myeloid Malignancies.
Lu JW, Hsieh MS, Liao HA, Yang YJ, Ho YJ, Lin LI. Lu JW, et al. Biomed Res Int. 2015;2015:641475. doi: 10.1155/2015/641475. Epub 2015 May 3. Biomed Res Int. 2015. PMID: 26064935 Free PMC article. Review. - Endothelial Loss of ETS1 Impairs Coronary Vascular Development and Leads to Ventricular Non-Compaction.
Wang L, Lin L, Qi H, Chen J, Grossfeld P. Wang L, et al. Circ Res. 2022 Aug 19;131(5):371-387. doi: 10.1161/CIRCRESAHA.121.319955. Epub 2022 Jul 27. Circ Res. 2022. PMID: 35894043 Free PMC article. - Analyses of Avascular Mutants Reveal Unique Transcriptomic Signature of Non-conventional Endothelial Cells.
Pak B, Schmitt CE, Choi W, Kim JD, Han O, Alsiö J, Jung DW, Williams DR, Coppieters W, Stainier DYR, Jin SW. Pak B, et al. Front Cell Dev Biol. 2020 Nov 23;8:589717. doi: 10.3389/fcell.2020.589717. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33330468 Free PMC article. - LSD1 promotes the egress of hematopoietic stem and progenitor cells into the bloodstream during the endothelial-to-hematopoietic transition.
Tamaoki J, Maeda H, Kobayashi I, Takeuchi M, Ohashi K, Gore A, Bonkhofer F, Patient R, Weinstein BM, Kobayashi M. Tamaoki J, et al. Dev Biol. 2023 Sep;501:92-103. doi: 10.1016/j.ydbio.2023.06.012. Epub 2023 Jun 22. Dev Biol. 2023. PMID: 37353106 Free PMC article.
References
- Baltzinger M, Mager-Heckel AM, Remy P. Xl erg: expression pattern and overexpression during development plead for a role in endothelial cell differentiation. Dev Dyn. 1999;216:420–33. - PubMed
- Bartel FO, Higuchi T, Spyropoulos DD. Mouse models in the study of the Ets family of transcription factors. Oncogene. 2000;19:6443–54. - PubMed
- Barton K, Muthusamy N, Fischer C, Ting CN, Walunas TL, Lanier LL, Leiden JM. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9:555–63. - PubMed
- Bories JC, Willerford DM, Grevin D, Davidson L, Camus A, Martin P, Stehelin D, Alt FW. Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature. 1995;377:635–8. - PubMed
- Brown LA, Rodaway AR, Schilling TF, Jowett T, Ingham PW, Patient RK, Sharrocks AD. Insights into early vasculogenesis revealed by expression of the ETS-domain transcription factor Fli-1 in wild-type and mutant zebrafish embryos. Mech Dev. 2000;90:237–52. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous