Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host - PubMed (original) (raw)
Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host
Justin L Sonnenburg et al. PLoS Biol. 2006 Nov.
Abstract
Probiotics are deliberately ingested preparations of live bacterial species that confer health benefits on the host. Many of these species are associated with the fermentation of dairy products. Despite their increasing use, the molecular details of the impact of various probiotic preparations on resident members of the gut microbiota and the host are generally lacking. To address this issue, we colonized germ-free mice with Bacteroides thetaiotaomicron, a prominent component of the adult human gut microbiota, and Bifidobacterium longum, a minor member but a commonly used probiotic. Simultaneous whole genome transcriptional profiling of both bacterial species in their gut habitat and of the intestinal epithelium, combined with mass-spectrometric analysis of habitat-associated carbohydrates, revealed that the presence of B. longum elicits an expansion in the diversity of polysaccharides targeted for degradation by B. thetaiotaomicron (e.g., mannose- and xylose-containing glycans), and induces host genes involved in innate immunity. Although the overall transcriptome expressed by B. thetaiotaomicron when it encounters B. longum in the cecum is dependent upon the genetic background of the mouse (as assessed by a mixed analysis of variance [ANOVA] model of co-colonization experiments performed in NMRI and C57BL/6J animals), B. thetaiotaomicron's expanded capacity to utilize polysaccharides occurs independently of host genotype, and is also observed with a fermented dairy product-associated strain, Lactobacillus casei. This gnotobiotic mouse model provides a controlled case study of how a resident symbiont and a probiotic species adapt their substrate utilization in response to one another, and illustrates both the generality and specificity of the relationship between a host, a component of its microbiota, and intentionally consumed microbial species.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
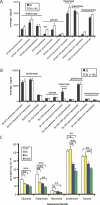
Figure 1. Impact of Co-colonization on B. thetaiotaomicron and B. longum Glycoside Hydrolase and Polysaccharide Lyase Gene Expression
(A) and (B) Bacterial glycoside hydrolase and polysaccharide lyase genes (rows) exhibiting differential expression in mono-associated versus co-colonized mice. Colors indicate the deviation of a gene's signal above (red) and below (green) its mean expression value across all ten samples. Thirty-three B. thetaiotaomicron enzymes and seven B. longum enzymes meet the selection criteria for differential expression described in Materials and Methods (FDR < 0.01). Fold-differences in average expression levels between the two colonization states are indicated in parenthesis for each gene. (C) and (D) Average aggregate signal intensity in the two colonization states for the seven groups of glycoside hydrolases/polysaccharide lyases that are common to B. thetaiotaomicron (C) and B. longum (D). The number of enzymes in each group is indicated in parenthesis. Values obtained from mono-associated mice are shown with black bars, and from co-colonized mice with white bars. See Tables S14 and S15 for a list of the B. thetaiotaomicron and B. longum genes, respectively. See Materials and Methods for a description of how “% maximum aggregate signal” was calculated. Mean values ± standard error (S.E.) of five samples are shown: an asterisk (*) indicates p < 0.05, and double asterisks (**) indicate p < 0.001 according to Student _t_-test.
Figure 2. Carbohydrate Utilization by B. thetaiotaomicron and B. longum in the Presence or Absence of One Another
(A) and (B) Average expression levels for five cecal samples are shown for genes encoding enzymes that shunt the indicated monosaccharides into the glycolytic or pentose phosphate pathways. Values obtained from mono-associated mice are shown with black bars, and from co-colonized mice with white bars. Significant differences in expression levels were determined using Student _t_-test; double asterisks (**) indicate p < 0.01, and triple asterisks (***) indicate p < 0.001. (C) GC-MS determination of monosaccharide composition in the ceca of germ-free (GF), B. thetaiotaomicron mono-associated (Bt), B. longum mono-associated (Bl), or B. thetaiotaomicron/B. longum co-colonized (Bt/Bl) mice. Mean values ± S.E. of biological triplicates are plotted. Significant changes in the abundance of a monosaccharide were identified using a one-way ANOVA followed by a Tukey post test; a single asterisk (*) indicates p < 0.05, double asterisks (**) indicate p < 0.01, and triple asterisks (***) indicate p < 0.001.
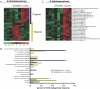
Figure 3. Changes in the B. thetaiotaomicron Transcriptome in Response to Co-colonization with B. animalis or L. casei
(A) B. thetaiotaomicron genes that show significantly increased expression levels upon co-colonization with either of the two fermented dairy product–associated strains are shown. Selection criteria: (1) a 1.2-fold or greater difference in expression using the 90% confidence bound of the fold-change; (2) difference in signal ≥ 25; (3) _p_-value ≤ 0.025 using Student _t_-test; (4) transcript called “present” in 66% or more of the GeneChips in the co-colonized group; and (5) a median FDR less than 0.05 based on 50 permutated comparisons. (B) COG-based functional classification of genes identified in (A). Thirty-five of 53 genes with higher expression in the presence of L. casei and 30 of 58 genes with higher expression in the presence of B. animalis could be assigned to COGs. Functional categories with significant over-representation in the dataset of differentially expressed genes, compared to their representation in the B. thetaiotaomicron genome, were determined using a hypergeometric distribution; an asterisk (*) indicates p < 0.01. (C) All B. thetaiotaomicron SusC/D paralogs and glycoside hydrolases/polysaccharide lyases that show differential expression between the three colonization states are up-regulated in the presence of L. casei. Fold-differences are indicated in parenthesis and reflect average expression across three cecal samples when B. thetaiotaomicron is co-colonized with L. casei, compared to their average expression in three cecal samples obtained from B. thetaiotaomicron mono-associated animals.
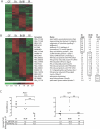
Figure 4. Gene Expression Changes in the Cecal Epithelium Induced by Colonization
(A) Cluster diagram from GeneChip studies of laser capture microdissected mouse cecal epithelium illustrates genes that meet the four criteria described in Materials and Methods including a 2-fold or greater increase in expression (using the 90% confidence bound of fold-change) in B. thetaiotaomicron mono-associated (Bt), B. longum mono-associated (Bl), or co-colonized ceca (Bt/Bl) compared with germ-free (GF) controls. (B) Genes involved in immuno-inflammatory responses that are up-regulated in at least one colonization state; fold-increase in expression in each colonization state is relative to the germ-free state. The expression pattern for Pap (Pancreatitis associated protein, a member of the C-type family of lectins known to bind terminal glycan structures in fungi and bacteria), and several other genes was confirmed using an independent GeneChip platform that queries a subset of genes in the mouse genome relevant to carbohydrate metabolism (Glyco-GeneChips; see Figure S9). (C) Real-time quantitative PCR assays of laser-captured microdissected cecal epithelial RNA isolated from individual animals colonized with B. thetaiotaomicron (Bt), B. longum (Bl), or co-colonized with both organisms (Bt/Bl) (n = 4 mice/group), using primers specific for Reg3γ or Isg15. Fold-changes are expressed relative to a reference RNA created by pooling equivalent masses of laser-captured microdissected cecal epithelial RNA from four germ-free animals. Average fold-changes for each group are indicated at the bottom, together with fold-changes derived from the GeneChip analysis. Significant differences in expression between groups for each gene were identified using a one-way ANOVA and Tukey post test; double asterisks (**) indicate p < 0.01, and triple asterisks (***) indicate p < 0.001.
Comment in
- In the gut's microbial community, one plus one equals many (effects).
Robinson R. Robinson R. PLoS Biol. 2006 Dec;4(12):e447. doi: 10.1371/journal.pbio.0040447. Epub 2006 Nov 28. PLoS Biol. 2006. PMID: 20076522 Free PMC article. No abstract available.
References
- Bengmark S, Martindale R. Prebiotics and synbiotics in clinical medicine. Nutr Clin Pract. 2005;20:244–261. - PubMed
- Molin G. Probiotics in foods not containing milk or milk constituents, with special reference to Lactobacillus plantarum 299v. Am J Clin Nutr. 2001;73:380S–385S. - PubMed
- Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347:911–920. - PubMed
- Metchnikoff É. In: The prolongation of life. Mitchell PC, translator. New York: Putnam; 1910. 343
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK030292/DK/NIDDK NIH HHS/United States
- DK30292/DK/NIDDK NIH HHS/United States
- GM62116/GM/NIGMS NIH HHS/United States
- U54 GM062116/GM/NIGMS NIH HHS/United States
- R37 DK030292/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases