Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3 - PubMed (original) (raw)
Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3
Peng Zhang et al. Nucleic Acids Res. 2006.
Abstract
Human MRG15 is a transcription factor that plays a vital role in embryonic development, cell proliferation and cellular senescence. It comprises a putative chromo domain in the N-terminal part that has been shown to participate in chromatin remodeling and transcription regulation. We report here the crystal structure of human MRG15 chromo domain at 2.2 A resolution. The MRG15 chromo domain consists of a beta-barrel and a long alpha-helix and assumes a structure more similar to the Drosophila MOF chromo barrel domain than the typical HP1/Pc chromo domains. The beta-barrel core contains a hydrophobic pocket formed by three conserved aromatic residues Tyr26, Tyr46 and Trp49 as a potential binding site for a modified residue of histone tail. However, the binding groove for the histone tail seen in the HP1/Pc chromo domains is pre-occupied by an extra beta-strand. In vitro binding assay results indicate that the MRG15 chromo domain can bind to methylated Lys36, but not methylated Lys4, Lys9 and Lys27 of histone H3. These data together suggest that the MRG15 chromo domain may function as an adaptor module which can bind to a modified histone H3 in a mode different from that of the HP1/Pc chromo domains.
Figures
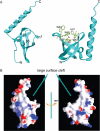
Figure 1
Structure of human MRG15 chromo domain. (A) Overall structure. Left panel: secondary structure elements. Right panel: structure of the potential binding pocket for a modified residue. Residues forming the pocket are shown with side chains. (B) Electrostatic surface of the MRG15 chromo domain. The β-barrel core and the C-terminal α-helix form a large surface cleft.
Figure 2
Comparison of the MRG15 chromo domain with representative chromo and chromo-like domains. (A) Sequence comparison of the chromo domains between MRG15 and its homologues in other species. Hs, Homo sapiens; Mm, Mus musculus; Dm, Drosophila melanogaster; Sp, Schizosaccharomyces pombe; Sc, Saccharomyces cerevisiae; At, Arabidopsis thaliana; and Ce, Caenorhabditis elegans. Strictly conserved residues are highlighted in shaded red boxes and conserved residues in open red boxes. The secondary structure of the MRG15 chromo domain is placed on top of the alignment. (B) Structure-based sequence alignment of the MRG15 chromo domain with representative chromo, Tudor and PWWP domains. The secondary structure for the first member of each group is placed on top of the alignment. Dm_MOF: the dMOF chromo barrel domain, PDB code 2BUD; Mm_MOF: the mouse MOF chromo barrel domain, PDB code 1WGS; Mm_HP1b: the mouse HP1β chromo domain, PDB code 1GUW; Dm_HP1: the Dm HP1 chromo domain, PDB code 1KNA; Dm_Pc: the Dm Pc chromo domain, PDB code 1PDQ; Sp_CLR4: the Sp CLR4 chromo domain, PDB code 1G6Z; Hs_SMN: the human SMN Tudor domain, PDB code 1G5V; Mm_53BP1: the mouse 53BP1 Tudor domain, PDB code 1XNI; Aa_NusG: the Aquifex aeolicus transcription factor NusG Tudor domain, PDB code 1M1G; Mm_Dnmt3b: the mouse Dnmt3b PWWP domain, PDB code 1KHC; Hs_HDGF: the human HDGF domain of hepatoma-derived growth factor (HDGF)-related protein, PDB code 1RIO; Sp_SPBC215: the Sp protein SPBC215 PWWP domain, PDB code 1H3Z; Mm_HRP: the PWWP domain of mouse HDGF-related protein 3, PDB code 1N27. The stars indicate conserved residues that form the hydrophobic pocket in the HP1/Pc chromo domains and the triangle indicates the residue that occupies in part the hydrophobic pocket. (C) Structural comparison of the MRG15 chromo domain with the dMOF and HP1 chromo domain, the HP1 chromo shadow domain, the SMN Tudor domain and the Dnmt3b PWWP domain. Residues forming the hydrophobic pocket are shown with side chains and the bound peptides in the HP1 chromo domain complex and the HP1 chromo shadow domain complex are shown in magenta. (D) Superposition of the MRG15 chromo domain (red), the dMOF chromo barrel domain (magenta), the HP1 chromo domain (yellow), the SMN Tudor domain (cyan) and the Dnmt3b PWWP domain (green). The bound histone H3 peptide in complex with the HP1 chromo domain is shown in blue.
Figure 3
In vitro binding assays showing that the MRG15 chromo domain can bind to methylated H3K36. (A) GST pull-down assays of the GST-fused MRG15 chromo domain (MRG15N) and the mouse HP1α with the calf thymus histone mixture. The protein samples were analyzed by SDS–PAGE with Coomassie blue staining. The results clearly show that the MRG15 chromo domain can bind to histone H3. (B) Western blot analysis of the GST pull-down samples of the MRG15 chromo domain (MRG15N), the mouse HP1α and the yeast Eaf3p chromo domain (Eaf3pN) with the calf thymus histone mixture. The mouse HP1α binds to H3K9me2 and H3K27me2. The yeast Eaf3p chromo domain binds to H3K36me2 and H3K4me3. The MRG15 chromo domain binds to H3K36me2. GST was used as a negative control. (C) Control experiments showing the specificity of the anti-H3K36me2/3 antibodies. The anti-H3K36me2 antibody has a high specificity with the H3K36me2 peptide and a weak cross-reaction with the H3K36me3 peptide, but no reaction with the unmethylated H3K36 peptide. The anti-H3K36me3 antibody recognizes only the H3K36me3 peptide. (D) In vitro binding assays of the N-terminal His-tagged MRG15 chromo domain with the H3K36me2 and H3K36me3 peptides. The yeast Eaf3p chromo domain (Eaf3pN) was used as a positive control. The results show that the MRG15 chromo domain can bind to both H3K36me2 and H3K36me3 peptides, but not the unmethylated H3K36 peptide.
Similar articles
- Molecular basis of the interaction of Saccharomyces cerevisiae Eaf3 chromo domain with methylated H3K36.
Sun B, Hong J, Zhang P, Dong X, Shen X, Lin D, Ding J. Sun B, et al. J Biol Chem. 2008 Dec 26;283(52):36504-12. doi: 10.1074/jbc.M806564200. Epub 2008 Nov 4. J Biol Chem. 2008. PMID: 18984594 Free PMC article. - Structural and biochemical studies on the chromo-barrel domain of male specific lethal 3 (MSL3) reveal a binding preference for mono- or dimethyllysine 20 on histone H4.
Moore SA, Ferhatoglu Y, Jia Y, Al-Jiab RA, Scott MJ. Moore SA, et al. J Biol Chem. 2010 Dec 24;285(52):40879-90. doi: 10.1074/jbc.M110.134312. Epub 2010 Oct 12. J Biol Chem. 2010. PMID: 20943666 Free PMC article. - Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain.
Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Bannister AJ, et al. Nature. 2001 Mar 1;410(6824):120-4. doi: 10.1038/35065138. Nature. 2001. PMID: 11242054 - Effector proteins for methylated histones: an expanding family.
Daniel JA, Pray-Grant MG, Grant PA. Daniel JA, et al. Cell Cycle. 2005 Jul;4(7):919-26. doi: 10.4161/cc.4.7.1824. Epub 2005 Jul 5. Cell Cycle. 2005. PMID: 15970672 Review. - Molecular biology of the chromo domain: an ancient chromatin module comes of age.
Eissenberg JC. Eissenberg JC. Gene. 2001 Sep 5;275(1):19-29. doi: 10.1016/s0378-1119(01)00628-x. Gene. 2001. PMID: 11574148 Review.
Cited by
- MRG15 promotes cell apoptosis through inhibition of mitophagy in hyperlipidemic acute pancreatitis.
Gu B, Yu W, Huang Z, Bai J, Liu S, Ren B, Wang P, Sun L, Wen J, Zheng Y, Tan P, Fu W. Gu B, et al. Apoptosis. 2024 Nov 2. doi: 10.1007/s10495-024-02034-4. Online ahead of print. Apoptosis. 2024. PMID: 39487311 - Genomic context-dependent histone H3K36 methylation by three Drosophila methyltransferases and implications for dedicated chromatin readers.
Jayakrishnan M, Havlová M, Veverka V, Regnard C, Becker PB. Jayakrishnan M, et al. Nucleic Acids Res. 2024 Jul 22;52(13):7627-7649. doi: 10.1093/nar/gkae449. Nucleic Acids Res. 2024. PMID: 38813825 Free PMC article. - Structural and functional insights into the epigenetic regulator MRG15.
Jiang N, Li YB, Jin JY, Guo JY, Ding QR, Meng D, Zhi XL. Jiang N, et al. Acta Pharmacol Sin. 2024 May;45(5):879-889. doi: 10.1038/s41401-023-01211-6. Epub 2024 Jan 8. Acta Pharmacol Sin. 2024. PMID: 38191914 Review. - Histone Readers and Their Roles in Cancer.
Wen H, Shi X. Wen H, et al. Cancer Treat Res. 2023;190:245-272. doi: 10.1007/978-3-031-45654-1_8. Cancer Treat Res. 2023. PMID: 38113004 Free PMC article. - Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Li H, Zhang Y, Wong KH, Zhang X, Chao WCH, He J. Dong S, et al. Cell Res. 2023 Oct;33(10):790-801. doi: 10.1038/s41422-023-00869-1. Epub 2023 Sep 4. Cell Res. 2023. PMID: 37666978 Free PMC article.
References
- Bertram M.J., Berube N.G., Hang-Swanson X., Ran Q., Leung J.K., Bryce S., Spurgers K., Bick R.J., Baldini A., Ning Y., et al. Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol. Cell. Biol. 1999;19:1479–1485. - PMC - PubMed
- Bertram M.J., Pereira-Smith O.M. Conservation of the MORF4 related gene family: identification of a new chromo domain subfamily and novel protein motif. Gene. 2001;266:111–121. - PubMed
- Marin I., Baker B.S. Origin and evolution of the regulatory gene male-specific lethal-3. Mol. Biol. Evol. 2000;17:1240–1250. - PubMed
- Pardo P.S., Leung J.K., Lucchesi J.C., Pereira-Smith O.M. MRG15, a novel chromodomain protein, is present in two distinct multiprotein complexes involved in transcriptional activation. J. Biol. Chem. 2002;277:50860–50866. - PubMed