Characterization of the short RNAs bound by the P19 suppressor of RNA silencing in mouse embryonic stem cells - PubMed (original) (raw)
. 2006 Dec;12(12):2092-102.
doi: 10.1261/rna.224606. Epub 2006 Oct 24.
Affiliations
- PMID: 17135486
- PMCID: PMC1664724
- DOI: 10.1261/rna.224606
Characterization of the short RNAs bound by the P19 suppressor of RNA silencing in mouse embryonic stem cells
J Mauro Calabrese et al. RNA. 2006 Dec.
Abstract
Studies of mammalian RNA interference (RNAi) have focused largely on the actions of microRNAs; however, in other organisms, endogenous short-interfering RNAs (siRNAs) are involved in silencing processes. To date, similar molecules have been difficult to characterize in mammalian cells. P19 is a plant suppressor of RNA silencing that binds with high affinity to siRNAs. Here, the short RNAs bound by P19 in mouse embryonic stem (ES) cells have been characterized. We show that P19 selectively immunoprecipitates endogenous short RNAs from ES cells. Cloning of immunoprecipitated RNA reveals a strong selection for short RNAs that are exact matches to ribosomal RNA (rRNA), with particular short rRNA species highly enriched in P19 immunoprecipitates. Complementary strands to the enriched rRNAs were not cloned, which was surprising because P19 was previously thought to bind only siRNAs. We show that P19 binds tightly to a noncanonical dsRNA substrate comprised of a short RNA annealed to a much longer partner, such that the double-stranded region between the two is 19 base pairs long. Binding to similar endogenous species might explain the association of P19 with short rRNAs in ES cells. Finally, we show that the P19-enriched rRNAs are not involved in canonical RNAi, as they exist in the absence of Dicer and do not function as post-transcriptional gene silencers. Our results support the previous observation that endogenous siRNAs are not abundant molecules in mouse ES cells.
Figures
FIGURE 1.
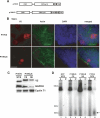
P19 binds endogenous short RNAs when expressed in ES cells. (A) P19 expression constructs used in this study. (B) Subcellular localization of P19V5 and P19NLS in ES cells (bar, 15 μm). (C) Western blots showing protein composition of P19-containing extracts. GAPDH (cytoplasmic) and Cyclin T1 (nuclear) serve as fractionation controls. (WCE) whole cell extract, (CE) cytoplasmic extract, (NE) nuclear extract. (D) P19 constructs bind short RNAs when expressed in ES cells. Cells transfected with GFP, P19V5, or P19NLS were lysed with either WCE or NE buffer, and immunoprecipitations were performed with αV5 Protein G agarose beads. Bound RNAs were 3′-end labeled with 5′-32P cytidine 3′, 5′-bis(phosphate) and resolved on a 12% denaturing polyacrylamide gel. The size markers correspond to a 10-bp DNA ladder. (Arrow) A ∼20-nt band observed in the P19 immunoprecipitations.
FIGURE 2.
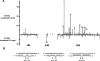
P19 enriches for particular short rRNA species when expressed in ES cells. (A) Specific short rRNAs are highly enriched in P19 immunoprecipitations compared with control supernatants. Shown is a scaled representation of all the short rRNAs cloned, aligned to bases 3900 to 13,000 of the 13,404-bp 45S pre-rRNA. Highlighted in bold along the X axis are the locations of the mature 18S, 5.8S, and 28S rRNA species relative to the full-length 45S pre-rRNA. Each gray bar represents one cloned short rRNA positioned directly above or below its matching sequence in the 45S pre-rRNA. Gray bars above the X axis were cloned from one or both of the immunoprecipitations, and those below the X axis were cloned from one or both of the supernatants. (B) Certain P19-enriched short rRNAs form partial double-stranded RNA structures with themselves. Shown are selected Mfold-predicted dsRNA structures of enriched rRNAs folded against each other.
FIGURE 3.
P19 binds with high affinity to RNAs containing 19- to 21-bp double-stranded regions with extended 5′- or 3′-single-stranded segments. (A) Base composition and secondary structure of short RNAs tested for binding to P19V5. In all cases, the strand in gray is short rRNA #3 from Fig. 2A. (B) Representative P19-binding assay. Shown is the RNA bound by P19V5–bead complexes after incubation with increasing concentrations of radiolabeled RNA (1, 5, 10, 50, and 100 nM). (C) Determination of the affinity of P19 for selected RNA species. Shown is the quantitation of a binding assay similar to that in B. The Kapp was determined by fitting the average data points to a fixed endpoint curve using KaleidaGraph data analysis software.
FIGURE 4.
Endogenous short rRNAs exist independently of Dicer. (A) P19 associates with short RNAs in the absence of Dicer. Dicer +/+ or −/− cells were transfected with either P19V5 or GFP as a negative control. Immunoprecipitations using V5 antibody were performed, and the associated RNA was 3′-end labeled and visualized as in Fig. 1D. (B) Similar RNA species associate with P19V5 in the presence or absence of Dicer. Shown is a short RNA Northern blot probing immunoprecipitated and supernatant RNA from A with probes complementary to RNA #3, miR295, or U6 snRNA.
Similar articles
- Studies of the interaction of the viral suppressor of RNA silencing protein p19 with small RNAs using fluorescence polarization.
Cheng J, Sagan SM, Jakubek ZJ, Pezacki JP. Cheng J, et al. Biochemistry. 2008 Aug 5;47(31):8130-8. doi: 10.1021/bi800401y. Epub 2008 Jul 3. Biochemistry. 2008. PMID: 18597480 - Enhanced specificity of the viral suppressor of RNA silencing protein p19 toward sequestering of human microRNA-122.
Cheng J, Danielson DC, Nasheri N, Singaravelu R, Pezacki JP. Cheng J, et al. Biochemistry. 2011 Sep 13;50(36):7745-55. doi: 10.1021/bi2008273. Epub 2011 Aug 17. Biochemistry. 2011. PMID: 21819044 - Characterization of silencing suppressor 2b of cucumber mosaic virus based on examination of its small RNA-binding abilities.
Goto K, Kobori T, Kosaka Y, Natsuaki T, Masuta C. Goto K, et al. Plant Cell Physiol. 2007 Jul;48(7):1050-60. doi: 10.1093/pcp/pcm074. Epub 2007 Jun 13. Plant Cell Physiol. 2007. PMID: 17567638 - Small RNAs derived from longer non-coding RNAs.
Röther S, Meister G. Röther S, et al. Biochimie. 2011 Nov;93(11):1905-15. doi: 10.1016/j.biochi.2011.07.032. Epub 2011 Aug 9. Biochimie. 2011. PMID: 21843590 Review. - Derivation and function of small interfering RNAs and microRNAs.
Cullen BR. Cullen BR. Virus Res. 2004 Jun 1;102(1):3-9. doi: 10.1016/j.virusres.2004.01.009. Virus Res. 2004. PMID: 15068874 Review.
Cited by
- Computational meta-analysis of ribosomal RNA fragments: potential targets and interaction mechanisms.
Guan L, Grigoriev A. Guan L, et al. Nucleic Acids Res. 2021 Apr 19;49(7):4085-4103. doi: 10.1093/nar/gkab190. Nucleic Acids Res. 2021. PMID: 33772581 Free PMC article. - RNA sequence analysis defines Dicer's role in mouse embryonic stem cells.
Calabrese JM, Seila AC, Yeo GW, Sharp PA. Calabrese JM, et al. Proc Natl Acad Sci U S A. 2007 Nov 13;104(46):18097-102. doi: 10.1073/pnas.0709193104. Epub 2007 Nov 7. Proc Natl Acad Sci U S A. 2007. PMID: 17989215 Free PMC article. - MicroRNAs and their potential involvement in HIV infection.
Sun G, Rossi JJ. Sun G, et al. Trends Pharmacol Sci. 2011 Nov;32(11):675-81. doi: 10.1016/j.tips.2011.07.003. Epub 2011 Aug 19. Trends Pharmacol Sci. 2011. PMID: 21862142 Free PMC article. Review. - Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs.
Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA. Leung AK, et al. Nat Struct Mol Biol. 2011 Feb;18(2):237-44. doi: 10.1038/nsmb.1991. Epub 2011 Jan 23. Nat Struct Mol Biol. 2011. PMID: 21258322 Free PMC article. - Recognition mechanism of siRNA by viral p19 suppressor of RNA silencing: a molecular dynamics study.
Xia Z, Zhu Z, Zhu J, Zhou R. Xia Z, et al. Biophys J. 2009 Mar 4;96(5):1761-9. doi: 10.1016/j.bpj.2008.11.047. Biophys J. 2009. PMID: 19254536 Free PMC article.
References
- Alvarez-Garcia, I., Miska, E.A. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. - PubMed
- Ambros, V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
- Ambros, V., Lee, R.C., Lavanway, A., Williams, P.T., Jewell, D. MicroRNAs and other tiny endogenous RNAs in C. elegans . Curr. Biol. 2003;13:807–818. - PubMed
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
- Baulcombe, D. RNA silencing in plants. Nature. 2004;431:356–363. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases