hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37 - PubMed (original) (raw)
hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37
Xiao-Bo Qiu et al. EMBO J. 2006.
Abstract
The 26S proteasome catalyzes the degradation of most proteins in mammalian cells. To better define its composition and associated regulatory proteins, we developed affinity methods to rapidly purify 26S proteasomes from mammalian cells. By this approach, we discovered a novel 46-kDa (407 residues) subunit of its 19S regulatory complex (previously termed ADRM1 or GP110). As its N-terminal half can be incorporated into the 26S proteasome and is homologous to Rpn13, a 156-residue subunit of the 19S complex in budding yeast, we renamed it human Rpn13 (hRpn13). The C-terminal half of hRpn13 binds directly to the proteasome-associated deubiquitinating enzyme, UCH37, and enhances its isopeptidase activity. Knockdown of hRpn13 in 293T cells increases the cellular levels of ubiquitin conjugates and decreases the degradation of short-lived proteins. Surprisingly, an overproduction of hRpn13 also reduced their degradation. Furthermore, transfection of the C-terminal half of hRpn13 slows proteolysis and induces cell death, probably by acting as a dominant-negative form. Thus in human 26S proteasomes, hRpn13 appears to be important for the binding of UCH37 to the 19S complex and for efficient proteolysis.
Figures
Figure 1
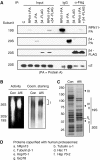
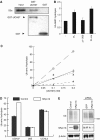
Affinity purification of mammalian proteasomes. (A) Immunoprecipitation of the affinity-tagged human proteasomes. 293T cells were transfected with mRPN11-protein-A, mβ4-protein A, mβ4-FLAG, or both mRPN11-protein-A and mβ4-FLAG. Following immunoprecipitation using an IgG or an anti-FLAG antibody, the subunits of both the 19S and 20S particles were detected by Western blot. Input represents 1% of the cell lysates for immunoprecipitation. (B) In-gel peptidase activity of the affinity purified proteasomes. Both the conventionally-purified (Con) and the affinity-purified (Affi) proteasomes were separated by native PAGE. Left, the peptidase activities of the proteasomes were assayed in the gel using suc-LLVY-amc as a substrate. Right, proteasomes were stained with Coomassie blue. (C) Analysis of affinity-purified proteasomes by SDS–PAGE. Both the conventionally purified (Con) and the affinity-purified (Affi) proteasomes were separated by SDS–PAGE and stained with Coomassie blue. (D) Mass spectrometric analysis of the proteins copurified with mammalian proteasomes. Protein samples for mass spectrometry were analyzed by MALDI-TOF using an Applied Biosystems Voyager DE-STR.
Figure 2
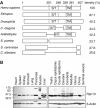
Rpn13 is evolutionally conserved in eukaryotes and is expressed in various mouse tissues. (A) hRpn13, especially its N-terminal half, is highly conserved through evolution. hRpn13 (GenBank accession no. NM 007002) and its homologs in several different organisms were aligned, and identities were calculated as indicated. A Ser/Thr-rich region (S/T) and a putative transmembrane (TM) domain in the C-terminal half are conserved in Rpn13 from multicellular organisms. (B) hRpn13 is expressed in various mouse tissues. Whole-tissue extracts were prepared and analyzed by Western blots.
Figure 3
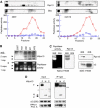
hRpn13 is a subunit of the 19S particle that associates with it through its N-terminal region. (A) Cosedimentation of hRpn13 with proteasomes during glycerol-gradient centrifugation. The lysates (3 mg proteins in 1 ml buffer) from 293T (left) or C2C12 (right) cells were loaded on a 10–40% of glycerol gradient, and centrifuged at 100 000 g (24 000 r.p.m., Beckman Rotor AH628) for 12 h. Peptidase activities from 26S (circle) and 20S (square) in each fraction were monitored by measuring the production of amc from the substrate suc-LLVY-amc in the absence and presence of 0.02% SDS, respectively. Distribution of S5a/Rpn10 (a 19S subunit) and hRpn13 was assayed in each fraction by Western blot. (B) hRpn13 associates with certain forms of proteasomes. Cell extracts (40 μg protein/well) were separated by native PAGE. In-gel peptidase activity was determined by incubating the gel with suc-LLVY-amc and then visualizing the active proteasomes under UV lights. (C) hRpn13 is present in the 19S particle. Left, the proteasomes purified by conventional methods were separated by native PAGE. In-gel peptidase activity was determined as in (B). Right, the proteasomes were separated by SDS–PAGE, and the α2 subunit of the 20S and hRpn13 were assayed by Western blot. (D) hRpn13 binds to proteasomes through its N-terminal region. 293T cells were cotransfected with mRPN11-protein-A and Myc/His6-tagged hRpn13 or its truncated forms. The cell lysates were immunoprecipitated using IgG beads. *stands for nonspecific bands ** heavy chain of IgG.
Figure 4
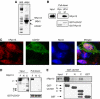
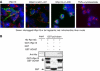
hRpn13 directly associates with UCH37. (A) hRpn13 co-immunoprecipitates with UCH37. 293T cells were transfected with hRpn13-Myc/His6, and incubated for 2 days. Following immunoprecipitation with an anti-Myc antibody, hRpn13-Myc and associated proteins were separated by SDS–PAGE, and stained with Coomassie blue. Conventionally purified 26S proteasomes were included in the gel as a reference. (B) hRpn13 directly binds to UCH37. Purified His-tagged hRpn13 and GST-fused UCH37 or Rpn12 were incubated for 1 h, and a pull-down assay was performed using GSH-beads. (C) UCH37 colocalizes with hRpn13. HeLa cells were transfected with Myc/His6-tagged UCH37, and the localization of the tagged UCH37 and endogenous hRpn13 was determined by immunostaining. (D) hRpn13 binds to UCH37 through its C-terminal region. 293T cells were either transfected with Myc-tagged hRpn13 (labeled as F) or with a similar construct encoding its N-terminal (N) or C-terminal half (C), and incubated for 2 days. Purified GST-UCH37 was incubated with the three cell lysates, and a pull-down assay was carried out using GSH-beads. (E) UCH37 binds to hRpn13 in vitro through its C-terminal half. Purified GST or GST-fused to the full-length (F), the N-terminal (N) or the C-terminal half (C) of UCH37 was incubated with purified recombinant His-tagged hRpn13. Pull-down assays using GSH-beads were then performed, and proteins were analyzed by Western blot.
Figure 5
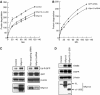
hRpn13 promotes isopeptidase activity of UCH37. (A) UCH37 binds to S5a in vitro. Purified GST or GST-fused UCH37 was incubated with purified recombinant His-tagged S5a for 1 h. Pull-down assays using GSH beads were then performed. The protein levels were analyzed by Western blot. (B) Recombinant hRpn13 stimulates the ubiquitin-amc (Ub-amc) hydrolysis by UCH37. In the presence of 0.5 μM of Ub-amc, 0.1 nM of UCH37 was incubated with 0 or 10 nM of the full-length hRpn13 (FL, from bacteria), the C-terminal half of hRpn13 (Δ1–200, from bacteria), or the full-length hRpn13 expressed in insect cells (FL-ins). Ub-amc hydrolysis (in arbitrary units) was determined during incubation for 30 min by monitoring the release of amc. (C) Unlike hRpn13, pure S5a/Rpn10 cannot promote the isopeptidase activity of UCH37 in vitro. UCH37 at indicated concentrations, was incubated with 0 (circle) or 10 nM of hRpn13 (square, expressed in insect cells), S5a (cross), or hRpn13 plus 10 nM of Ub-aldehyde (triangle). Ub-amc hydrolysis (in arbitrary units) was determined as in (B). (D) hRpn13 does not increase the isopeptidase activity of the 26S proteasome or UCHL3. Ub-amc was incubated (as in 5B) without (control) or with 10 nM of purified hRpn13 in the presence of UCH37 (0.1 nM), the 26S proteasome (0.24 μg/ml, about 0.1 nM), or UCHL3 (0.01 nM). Ub-amc hydrolysis (in arbitrary units) was determined as in (B). (E) hRpn13 decreases the levels of ubiquitin conjugates in cells. Left, 293T cells were transfected with an empty vector or Myc/His6-tagged hRpn13. Right, 293T cells were transfected with pBS/U6/GFP (GFP siRNA) or pBS/U6/hRpn13 (hRpn13 siRNA). The cells were incubated for 2 days after transfection, and the contents of hRpn13, β-actin, and large ubiquitin conjugates (molecular weights >191 kDa) were assayed by Western blot.
Figure 6
hRpn13 content influences rates of degradation of short-lived proteins in 293T cells. (A) Overexpression of hRpn13 or its C-terminal region reduces the degradation of short-lived proteins. 293T cells were transfected with an empty vector, hRpn13 or its C-terminal region (hRpn13-Δ1–200). After 2 days, the rate of degradation of cellular proteins was determined by pulse-chase analysis using [3H]tyrosine. A similar small reduction in degradation was seen in at least three experiments. (B) Decreasing the levels of hRpn13 also slows the degradation of short-lived proteins. 293T cells were transfected with pBS/U6/GFP (GFP siRNA) or pBS/U6/hRpn13 (hRpn13 siRNA). Then, degradation of short-lived proteins was determined as in (A). (C) Overexpression or knockdown of hRpn13 reduces the degradation of the model N-end rule substrate, Ub-R-GFP. Left, 293T cells were cotransfected with Ub-R-GFP, GFP, and hRpn13 (or empty vector). Right, 293T cells were cotransfected with Ub-R-GFP, GFP, and siRNA oligos (100 nM) for hRpn13 (or nontargeting siRNA oligos). The cells were incubated for 3 days after transfection, and the protein levels were analyzed by Western blot. (D) Degradation of the natural proteasomal substrate, ErbB3, is reduced by overexpression of hRpn13 or its C-terminal half. 293T cells were cotransfected with ErbB3, EGFR, and hRpn13 (or hRpn13-Δ1–200 or empty vector), and incubated for 2 days after transfection. Protein levels were analyzed by Western blot.
Figure 7
Expression of the C-terminal half (but not the N-terminal half) of hRpn13 induces cell death. (A) HeLa cells were transfected with Myc/His6-tagged hRpn13, hRpn13-Δ201–407, or hRpn13-Δ1–200. After 1 day, hRpn13 and its fragments were visualized by immunofluorescence staining with an anti-His6 antibody, mitochondria with MitroTracker Red, and nuclei with DAPI. The apoptosis-like features, the condensation of nuclei and the severe aggregation of mitochondria, were observed in more than 90% cells (about 50 cells examined) that were transfected with hRpn13-Δ1–200. For comparison with apoptotic cell death, the cells were treated with 20 ng/ml of TNFα and 1 μM of cycloheximide for 4 h. (B) The C-terminal half of hRpn13 does not bind to full-length hRpn13, though it binds to UCH37. The purified His-hRpn13-Δ1–200 was incubated with GST, GST-hRpn13 or GST-UCH37 for 1 h. Pull-down assays were performed using glutathione beads, and the protein levels were determined by Western blot. Thus, the C-terminal half of hRpn13 causes cell death, probably by blocking UCH37 binding to the proteasome.
Similar articles
- A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes.
Hamazaki J, Iemura S, Natsume T, Yashiroda H, Tanaka K, Murata S. Hamazaki J, et al. EMBO J. 2006 Oct 4;25(19):4524-36. doi: 10.1038/sj.emboj.7601338. Epub 2006 Sep 21. EMBO J. 2006. PMID: 16990800 Free PMC article. - Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1.
Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, Cohen RE. Yao T, et al. Nat Cell Biol. 2006 Sep;8(9):994-1002. doi: 10.1038/ncb1460. Epub 2006 Aug 13. Nat Cell Biol. 2006. PMID: 16906146 - Structure of the Rpn13-Rpn2 complex provides insights for Rpn13 and Uch37 as anticancer targets.
Lu X, Nowicka U, Sridharan V, Liu F, Randles L, Hymel D, Dyba M, Tarasov SG, Tarasova NI, Zhao XZ, Hamazaki J, Murata S, Burke TR Jr, Walters KJ. Lu X, et al. Nat Commun. 2017 Jun 9;8:15540. doi: 10.1038/ncomms15540. Nat Commun. 2017. PMID: 28598414 Free PMC article. - Deubiquitinating enzymes are IN/(trinsic to proteasome function).
Guterman A, Glickman MH. Guterman A, et al. Curr Protein Pept Sci. 2004 Jun;5(3):201-11. doi: 10.2174/1389203043379756. Curr Protein Pept Sci. 2004. PMID: 15188770 Review. - New insights into the role of the ubiquitin-proteasome pathway in the regulation of apoptosis.
Liu CH, Goldberg AL, Qiu XB. Liu CH, et al. Chang Gung Med J. 2007 Nov-Dec;30(6):469-79. Chang Gung Med J. 2007. PMID: 18350730 Review.
Cited by
- Regulation of NF-kappaB activity and inducible nitric oxide synthase by regulatory particle non-ATPase subunit 13 (Rpn13).
Mazumdar T, Gorgun FM, Sha Y, Tyryshkin A, Zeng S, Hartmann-Petersen R, Jørgensen JP, Hendil KB, Eissa NT. Mazumdar T, et al. Proc Natl Acad Sci U S A. 2010 Aug 3;107(31):13854-9. doi: 10.1073/pnas.0913495107. Epub 2010 Jul 15. Proc Natl Acad Sci U S A. 2010. PMID: 20634424 Free PMC article. - Design principles that protect the proteasome from self-destruction.
Singh Gautam AK, Yu H, Yellman C, Elcock AH, Matouschek A. Singh Gautam AK, et al. Protein Sci. 2022 Mar;31(3):556-567. doi: 10.1002/pro.4251. Epub 2021 Dec 16. Protein Sci. 2022. PMID: 34878680 Free PMC article. - Small-Molecule Inhibitors of the Proteasome's Regulatory Particle.
Muli CS, Tian W, Trader DJ. Muli CS, et al. Chembiochem. 2019 Jul 15;20(14):1739-1753. doi: 10.1002/cbic.201900017. Epub 2019 May 24. Chembiochem. 2019. PMID: 30740849 Free PMC article. Review. - Autoregulation of the 26S proteasome by in situ ubiquitination.
Jacobson AD, MacFadden A, Wu Z, Peng J, Liu CW. Jacobson AD, et al. Mol Biol Cell. 2014 Jun 15;25(12):1824-35. doi: 10.1091/mbc.E13-10-0585. Epub 2014 Apr 17. Mol Biol Cell. 2014. PMID: 24743594 Free PMC article. - Small-Molecule Inhibitors Targeting Proteasome-Associated Deubiquitinases.
Moon S, Muniyappan S, Lee SB, Lee BH. Moon S, et al. Int J Mol Sci. 2021 Jun 9;22(12):6213. doi: 10.3390/ijms22126213. Int J Mol Sci. 2021. PMID: 34207520 Free PMC article. Review.
References
- Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG (2000) Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol 18: 538–543 - PubMed
- Glickman MH, Ciechanover A (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82: 373–428 - PubMed
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94: 615–623 - PubMed
- Goldberg AL (2003) Protein degradation and protection against misfolded or damaged proteins. Nature 426: 895–899 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials