Editor meets silencer: crosstalk between RNA editing and RNA interference - PubMed (original) (raw)
Review
Editor meets silencer: crosstalk between RNA editing and RNA interference
Kazuko Nishikura. Nat Rev Mol Cell Biol. 2006 Dec.
Abstract
The most prevalent type of RNA editing is mediated by ADAR (adenosine deaminase acting on RNA) enzymes, which convert adenosines to inosines (a process known as A-->I RNA editing) in double-stranded (ds)RNA substrates. A-->I RNA editing was long thought to affect only selected transcripts by altering the proteins they encode. However, genome-wide screening has revealed numerous editing sites within inverted Alu repeats in introns and untranslated regions. Also, recent evidence indicates that A-->I RNA editing crosstalks with RNA-interference pathways, which, like A-->I RNA editing, involve dsRNAs. A-->I RNA editing therefore seems to have additional functions, including the regulation of retrotransposons and gene silencing, which adds a new urgency to the challenges of fully understanding ADAR functions.
Conflict of interest statement
Competing interests statement: The author declares no competing financial interests.
Figures
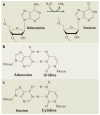
Figure 1. Deamination of adenosine to inosine by ADAR
a | A hydrolytic deamination reaction converts adenosine to inosine. b | Adenosine base-pairs with uridine. c | By contrast, inosine base-pairs, as if it were guanosine, in a Watson–Crick-bonding configuration with cytidine.
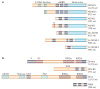
Figure 2. Types of dsRBD-containing protein: ADAR-family proteins and proteins that are required for miRNA biogenesis
a | Three human ADAR (adenosine deaminase acting on RNA)-family members (ADAR1–3), Drosophila melanogaster (Dm) ADAR and two Caenorhabditis elegans (Ce) proteins, ADAR-1 and ADAR-2, share common functional domains: 2 or 3 repeats of the dsRBD and a catalytic deaminase domain. Certain structural features, such as Z-DNA-binding domains and the Arg-rich (R) domain, are unique to particular ADAR members. Binding of ADAR to double-stranded (ds)RNA substrates is mediated through dsRBDs, whereas Z-DNA-binding domains might increase the affinity of ADAR1L specifically for short dsRNAs such as siRNAs. Binding of the R domain to single-stranded RNAs has been reported, but its biological significance is currently unknown. Two ADAR1 translation products, the isoforms ADAR1L and ADAR1S, result from transcription from different promoters followed by alternative splicing. This leads to translation initiation from the upstream or downstream Met codon. b | Drosha and Dicer, two RNase III endonuclease family members, are essential for miRNA biogenesis. Drosha and Dicer, as well as cofactors DGCR8 and TRBP, contain one or more dsRBDs. In addition to the catalytic domain RIIID, which is responsible for the RNase III endonucleolytic reaction, unique functional domains, such as the Pro-rich (P) and Arg–Ser-rich (RS) domains, are present in Drosha. By contrast, the DEAD-box RNA helicase, DUF and PAZ domains are present in Dicer. The PAZ domain binds to the 3′ end of miRNAs, whereas the precise role of the DEAD-box RNA helicase domain is unknown. The function of the DUF domain is also unknown. The WW motif of DGCR8 is likely to be involved in protein interactions. Both ADARs and the proteins involved in the miRNA biogenesis pathway bind their dsRNA substrates through dsRBDs. The interaction between dsRNA and dsRBD is not RNA-sequence specific. Therefore, adenosine to inosine (A→I) editing and RNA-interference mechanisms might compete for a common dsRNA substrate, such as primary transcript miRNA (FIGS 6,7). aa, amino acids.
Figure 3. Functional changes by A→I RNA editing of coding sequences
a |
l
-glutamate is the predominant excitatory neurotransmitter in vertebrate nervous systems, and the glutamate receptor (GluR) has been implicated in neuronal plasticity and higher functions such as memory and learning. Adenosine to inosine (A→I) RNA editing of the Gln/Arg (Q/R) site leads to the replacement of a Gln by an Arg residue,. Ion-channel receptors that contain the edited GluR2 subunit are impermeable to Ca2+, whereas channels that lack the edited subunit permit influx of Ca2+. Q/R-site editing also regulates the tetramerization and intracellular trafficking of the receptor protein. b | Serotonin receptors have important roles in physiological and behavioural processes such as circadian rhythms, emotional control and feeding behaviour,. G-protein-coupling functions of serotonin (5-HT) receptor-2C (5-HT2CR) are dramatically reduced by A→I RNA editing that occurs at five sites (A, B, C, D and E sites). For example, the potency of the agonist-stimulated G-protein-coupling activity of the fully edited receptor isoform (Val-Gly-Val) is reduced by 20-fold compared with the unedited receptor isoform (Ile-Asn-Ile),. The fold-back double-stranded (ds)RNA structure, which consists of short dsRNA regions, bulges and loops, is formed because of partial complementarity of the exon and intronic editing-site complementary sequence (ECS; which is essential for editing). The thick dark-blue line represents the exon, and the thin dark-blue line represents the intron. Certain sites are exclusively edited only by ADAR1 (adenosine deaminase acting on RNA-1) or ADAR2; ADAR2 edits exclusively the Q/R site of GluR2 subunit and the D site of 5-HT2CR, whereas ADAR1 selectively edits the A and B sites of 5-HT2CR. The molecular mechanism that underlies the editing-site selectivity is not yet completely understood. However, the secondary structure in the fold-back dsRNA substrates, as well as functional interactions between two monomers of ADAR1 or ADAR2, might dictate editing-site selectivity. Several intronic editing sites that have been detected in GluR2 and 5-HT2CR dsRNAs are not shown.
Figure 4. Extensive A→I RNA editing of non-coding repeat sequences
a | A typical alignment of genomic and expressed sequence tag (EST) cDNA sequences is shown. Adenosine to inosine (A→I) RNA editing sites that have been identified as A→ guanosine (G) changes are marked by arrows. b | Detection of A→I RNA editing sites (arrows) in intron 2 of the cyclin CNNM3 pre-mRNA. Both bioinformatics screening and PCR after reverse transcription of RNA (RT-PCR) experiments have identified numerous extensively edited sites (some sites are 80–90% edited) in the double-stranded (ds)RNA structure that contains two inversely oriented Alu-subfamily members, AluSg+ and _AluJb_− (REF. 76). c | Scarcity of an intermolecular RNA duplex with sense and antisense transcript pair. Co-expression of CNNM3 pre-mRNAs and antisense transcripts in NT2-N neurons was discovered while analysing RT-PCR products. As with CNNM3 pre-mRNA, extensive A→I RNA editing of these antisense transcripts was limited to the Alu-repeat sequences (shown in red and green), which indicated that sense and antisense strand RNAs formed two separate intramolecular dsRNAs instead of a completely complementary, long intermolecular dsRNA. Together, it seems that in vivo formation of intermolecular RNA duplexes of sense and antisense transcripts is very rare, if it occurs at all.
Figure 5. Possible regulatory functions for non-coding RNA editing
a | Extensive adenosine to inosine (A→I) editing of an RNA-duplex structure that consists of inverted Alu or LINE repeats. The inverted Alu or LINE repeats in introns and untranslated regions (UTRs) form intramolecular RNA duplexes genome wide, which are then subjected to A→I RNA editing by ADAR (adenosine deaminase acting on RNA). b | An inosine is interpreted by the splicing machinery as a guanosine. Therefore, splice sites might be created or deleted due to A→I editing of intronic Alu fold-back double-stranded (ds)RNAs, leading to the inclusion or exclusion of Alu exons. c | A→I editing of a SINE fold-back dsRNA present in the 3′ UTR of CTN-RNA and its binding to p54nrb might be involved in the regulatory mechanism that retains this RNA in nuclear speckles. When cells are placed under stress, CTN-RNA is cleaved and de novo polyadenylated at an alternative site to release the protein-coding Cat2 mRNA, which is then translated into cationic amino-acid transporter-2 protein. The factors involved in the cleavage and de novo polyadenylation mechanisms are unknown. d | Tudor staphylococcal nuclease (Tudor-SN), an RNA-induced silencing complex (RISC)-associated component that lacks an assigned function in the RNA interference (RNAi) mechanism, has recently been identified as a potential inosine-containing dsRNA (I-dsRNA)-specific ribonuclease. A→I editing of pre-mRNAs containing Alu or LINE fold-back dsRNA structures might be degraded by Tudor-SN, which, in turn, might control the expression levels of genes harbouring repeat sequences. e | The possibility that A→I RNA editing is involved in the heterochromatic silencing mechanism has been indicated by findings of Vigilin–ADAR1 complex formation and binding of Vigilin to inosine-containing RNAs. Vigilin is an RNA-binding protein localized both in the nucleus and cytoplasm. The Drosophila melanogaster homologue of Vigilin, DDP1, has been known to have a role in heterochromatic gene silencing. The heterochromatic silencing process modifies the chromatin structure through various mechanisms, including histone H3 Lys9 methylation (H3K9me) and HP1 binding, which might eventually lead to methylation of cytosines in DNA (see recent reviews on heterochromatic silencing86,87,91). HP1, heterochromatin protein-1; RHA, RNA helicase A. f | In somatic cells and tissues, A→I editing of Alu or LINE fold-back dsRNAs might suppress the generation of rasiRNAs and therefore RNAi-mediated silencing in trans of genes that harbour the Alu or LINE sequence in UTRs. In mouse oocytes, rasiRNAs are generated, possibly due to the absence of A→I editing. Modified with permission from REF. 112 © (2004) MacMillan Magazines Ltd.
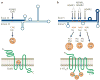
Figure 6. Interaction between RNA editing and RNA-interference pathways
Two ways of interaction between RNA editing and RNA-interference pathways have been proposed. a | The introduction of many inosine·uridine (I·U) mismatched base pairs and the alteration of the double-stranded (ds)RNA structure by ADAR (adenosine deaminase acting on RNA) leads to the generation of fewer siRNAs by Dicer, because such dsRNAs that contain many I·U mismatched base pairs become resistant to Dicer cleavage. b | Also, a fraction of already processed siRNAs might be sequestered by certain ADAR-gene-family members, reducing the effective siRNA concentration. For example, cytoplasmic ADAR1L binds siRNA tightly. Gene silencing by siRNA is significantly more effective in the absence of ADAR1, which indicates that ADAR1L is a cellular factor that limits siRNA potency in mammalian cells by decreasing the effective siRNA concentration and its incorporation into the RNA-induced silencing complex (RISC).
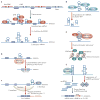
Figure 7. Regulation of microRNA processing and expression by RNA editing
a | Adenosine to inosine (A→I) editing sites of pri-miRNA-142. The region to be processed into mature sense and antisense strand miRNA-142 (5p and 3p, respectively) is highlighted in green. Five major editing sites are indicated by an A in red. The 5′ end of the mature miRNA-142-5p sequence is numbered as +1. Editing of the +4 and the +5 sites inhibits cleavage by the Drosha–DGCR8 complex. Modified with permission from REF. 19 © (2006) MacMillan Magazines Ltd. b | The Drosha-DGCR8 complex cleaves pri-miRNAs in the nucleus, producing ∼70-nucleotide pre-miRNA intermediates, which are exported by exportin-5 and RanGTP into the cytoplasm. The Dicer–TRBP complex executes the second cleavage, generating mature miRNAs. c | Drosha cleavage of pri- to pre-miRNA is suppressed by A→I editing of certain sites, such as the +4 and +5 sites of pri-miRNA-142. Also, highly edited pri-miRNA-142 is degraded by Tudor staphylococcal nuclease (Tudor-SN). d | A→I editing of certain sites might lead to inhibition of the Dicer–TRBP cleavage step. e | Editing of pri-miRNAs at certain sites might lead to the expression of ‘edited mature miRNAs’ (for example, Kaposi-sarcoma-associated virus miRNA, miRNA-K12-10b).
Comment in
- miRNA editing--we should have inosine this coming.
Habig JW, Dale T, Bass BL. Habig JW, et al. Mol Cell. 2007 Mar 23;25(6):792-3. doi: 10.1016/j.molcel.2007.03.010. Mol Cell. 2007. PMID: 17386255 Free PMC article.
Similar articles
- All I's on the RADAR: role of ADAR in gene regulation.
Shevchenko G, Morris KV. Shevchenko G, et al. FEBS Lett. 2018 Sep;592(17):2860-2873. doi: 10.1002/1873-3468.13093. Epub 2018 May 25. FEBS Lett. 2018. PMID: 29770436 Free PMC article. Review. - A-to-I editing of protein coding and noncoding RNAs.
Mallela A, Nishikura K. Mallela A, et al. Crit Rev Biochem Mol Biol. 2012 Nov-Dec;47(6):493-501. doi: 10.3109/10409238.2012.714350. Epub 2012 Sep 18. Crit Rev Biochem Mol Biol. 2012. PMID: 22988838 Review. - Noncoding regions of C. elegans mRNA undergo selective adenosine to inosine deamination and contain a small number of editing sites per transcript.
Wheeler EC, Washburn MC, Major F, Rusch DB, Hundley HA. Wheeler EC, et al. RNA Biol. 2015;12(2):162-74. doi: 10.1080/15476286.2015.1017220. RNA Biol. 2015. PMID: 25826568 Free PMC article. - RNA editing by mammalian ADARs.
Hogg M, Paro S, Keegan LP, O'Connell MA. Hogg M, et al. Adv Genet. 2011;73:87-120. doi: 10.1016/B978-0-12-380860-8.00003-3. Adv Genet. 2011. PMID: 21310295 Review. - In cancer, A-to-I RNA editing can be the driver, the passenger, or the mechanic.
Ganem NS, Ben-Asher N, Lamm AT. Ganem NS, et al. Drug Resist Updat. 2017 May;32:16-22. doi: 10.1016/j.drup.2017.09.001. Epub 2017 Oct 4. Drug Resist Updat. 2017. PMID: 29145975 Review.
Cited by
- Emerging Role of Environmental Epitranscriptomics and RNA Modifications in Parkinson's Disease.
Gionco JT, Bernstein AI. Gionco JT, et al. J Parkinsons Dis. 2024;14(4):643-656. doi: 10.3233/JPD-230457. J Parkinsons Dis. 2024. PMID: 38578904 Free PMC article. Review. - RNA modifications in cellular metabolism: implications for metabolism-targeted therapy and immunotherapy.
Liu WW, Zheng SQ, Li T, Fei YF, Wang C, Zhang S, Wang F, Jiang GM, Wang H. Liu WW, et al. Signal Transduct Target Ther. 2024 Mar 27;9(1):70. doi: 10.1038/s41392-024-01777-5. Signal Transduct Target Ther. 2024. PMID: 38531882 Free PMC article. Review. - Harnessing ADAR-Mediated Site-Specific RNA Editing in Immune-Related Disease: Prediction and Therapeutic Implications.
Weng S, Yang X, Yu N, Wang PC, Xiong S, Ruan H. Weng S, et al. Int J Mol Sci. 2023 Dec 26;25(1):351. doi: 10.3390/ijms25010351. Int J Mol Sci. 2023. PMID: 38203521 Free PMC article. Review. - Withania somnifera mitochondrial atp4 gene editing alters the ATP synthase b subunit, independent of salt stress.
Ramadan AM, Al-Ghamdi KM, Alghamdi AJ, Amer M, Ibrahim MIM, Atef A. Ramadan AM, et al. Saudi J Biol Sci. 2023 Nov;30(11):103817. doi: 10.1016/j.sjbs.2023.103817. Epub 2023 Sep 28. Saudi J Biol Sci. 2023. PMID: 37841665 Free PMC article. - L-GIREMI uncovers RNA editing sites in long-read RNA-seq.
Liu Z, Quinones-Valdez G, Fu T, Huang E, Choudhury M, Reese F, Mortazavi A, Xiao X. Liu Z, et al. Genome Biol. 2023 Jul 20;24(1):171. doi: 10.1186/s13059-023-03012-w. Genome Biol. 2023. PMID: 37474948 Free PMC article.
References
- Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. - PubMed
- Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu Rev Genet. 2000;34:499–531. - PubMed
- Valente L, Nishikura K. ADAR gene family and A-to-I RNA editing: diverse roles in posttranscriptional gene regulation. Prog Nucleic Acid Res Mol Biol. 2005;79:299–338. - PubMed
- An up-to-date review on A→I editing and ADAR genes.
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM040536/GM/NIGMS NIH HHS/United States
- R01 HL070045/HL/NHLBI NIH HHS/United States
- R01 GM040536-15/GM/NIGMS NIH HHS/United States
- P01 CA072765/CA/NCI NIH HHS/United States
- P01 CA072765-050002/CA/NCI NIH HHS/United States
- R01 HL070045-04/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials