Chimeric DNA methyltransferases target DNA methylation to specific DNA sequences and repress expression of target genes - PubMed (original) (raw)
Chimeric DNA methyltransferases target DNA methylation to specific DNA sequences and repress expression of target genes
Fuyang Li et al. Nucleic Acids Res. 2007.
Abstract
Gene silencing by targeted DNA methylation has potential applications in basic research and therapy. To establish targeted methylation in human cell lines, the catalytic domains (CDs) of mouse Dnmt3a and Dnmt3b DNA methyltransferases (MTases) were fused to different DNA binding domains (DBD) of GAL4 and an engineered Cys2His2 zinc finger domain. We demonstrated that (i) Dense DNA methylation can be targeted to specific regions in gene promoters using chimeric DNA MTases. (ii) Site-specific methylation leads to repression of genes controlled by various cellular or viral promoters. (iii) Mutations affecting any of the DBD, MTase or target DNA sequences reduce targeted methylation and gene silencing. (iv) Targeted DNA methylation is effective in repressing Herpes Simplex Virus type 1 (HSV-1) infection in cell culture with the viral titer reduced by at least 18-fold in the presence of an MTase fused to an engineered zinc finger DBD, which binds a single site in the promoter of HSV-1 gene IE175k. In short, we show here that it is possible to direct DNA MTase activity to predetermined sites in DNA, achieve targeted gene silencing in mammalian cell lines and interfere with HSV-1 propagation.
Figures
Figure 1
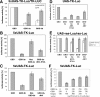
Targeted methylation at the 5×UAS-TK promoter region in HEK293T cells analyzed by bisulfite sequencing. (A) Schematic representation of the reporter constructs encoding the TK promoter fused to the luciferase firefly gene constructs as well as the constructs coding for targeted DBD-MTase fusion proteins. Targeted promoters contain one or five copies of the GAL4 binding sites (UAS). (B–D) Bisulfite sequencing of the promoter regions of the reporter constructs. Each line represents one individual clone sequenced. The empty squares denote for unmethylated CG sites, the filled ones for methylated sites. The location of the CG sites (1–26) within the 5×UAS-TK promoter region is schematically shown in (B). (B) Co-transfection of the 5×UAS-TK-Luc reporter construct with GBD only. (C) Co-transfection of 5×UAS-TK-Luc with GBD-3a or GBD-3b. The totally unmethylated clones #11 and 12 for 5×UAS-TK/GBD-3a and #9 and 10 for 5×UAS-TK/GBD-3b probably were PCR amplified from plasmids that were not taken up by the cells during the transfection procedure. (D) Co-transfection of 5×UAS-TK-Luc with the mutGBD-3a, mutGBD-3b, GBD-3a-ANV/E74A or GBD-3b-ANV variants. The mutGBD-3a and 3b variants carry an inactivated GBD domain. The MTase domain of the GBD-3a-ANV/E74A and GBD-3b-ANV is inactivated by mutations. (E) DNA methylation within the luciferase gene ∼1 kb downstream of the TK promoter after co-transfecion of the 5×UAS-TK-Luc reporter gene with GBD-3a and GBD-3b.
Figure 2
Specific repression of the luciferase reporter gene by GBD-MTase fusion constructs. (A) GBD-3a, GBD-3b and GBD expression constructs were co-transfected with firefly luciferase reporter plasmids 5×UAS-TK-Luc or TK-Luc into HEK293T cells and the reporter gene activity determined. (B) Co-transfection of the 5×UAS-TK-Luc reporter plasmid with GBD, GBD-3a, mutGBD-3a and GBD-3a-ANV/E74A expression vectors into HEK293T cells. (C) Co-transfection of the 5×UAS-TK-Luc reporter plasmid with GBD, GBD-3b, mutGBD-3b and GBD-3b-ANV expression vectors into HEK293T cells. (D) Co-transfection of the UAS-TK-Luc reporter plasmid with GBD, GBD-3a, GBD-3a-ANV/E74A, GBD-3b and GBD-3b-ANV into HEK293T cells. (E) Co-transfection of UAS-ras-Luc and ras-Luc reporter plasmids with the GBD, GBD-3a, GBD-3a-ANV/E74A, GBD-3b and GBD-3b-ANV into HEK293T cells. In each case the error bars give the standard deviations of at least three independent experiments. (F) Competition experiment of GBD-3a and GBD. To investigate the targeting function of the Zinc finger, GBD-3a or GBD-3b were co-transfected with the 5×UAS-TK-Luc reporter plasmid construct together with increasing amounts of a construct that expresses GBD. The total amount of DNA for transfection was normalized by empty vector. The numbers below the axis indicate the amount of DNA of each construct (ng/well in 24-well plate) used in each transfection.
Figure 3
Targeted methylation of the IE175k promoter by 6F6-3a or B1-3a as analyzed by bisulfite sequencing. (A) Schematic representation of the reporter constructs comprising the wild-type IE175k promoter (IE175) or the IE175k promoter with deleted B1/6F6 site (IE175mut) fused to firefly luciferase gene and the constructs encoding targeted methyltransferase fusion proteins B1-3a and 6F6-3a. (B–D) DNA Methylation analysis of the IE175k promoter after co-transfection with MTase expression constructs into HEK293T cells. Each line represents an individual clone. The empty squares represent unmethylated CG sites, whereas the filled ones methylated sites. In (B) the IE175-Luc reporter gene is co-transfected with 6F6. In (C) IE175-Luc is co-transfected with 6F6-3a or B1-3a and in (D) the IE175mut-Luc construct is co-transfected with 6F6-3a or B1-3a. The B1/6F6 binding site is marked in panel B. (E) DNA methylation within the luciferase gene ∼1 kb downstream of the target site (downstream site) in IE175-Luc and within the CMV promoter of the Renilla luciferase gene that was used for internal normalization in the presence of 6F6-3a and B1-3a.
Figure 4
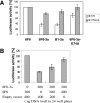
Targeted repression of the IE175k promoter by synthetic zinc-finger proteins fused to the CD of Dnmt3a. (A) The IE175-Luc or IE175mut-Luc reporter plasmids were co-transfected with 6F6, 6F6-3a, B1-3a or 6F6-3a-E74A into HEK293T cells. The IE175mut promoter does not contain the binding site for the engineered zinc finger protein B1/6F6. The Dnmt3a-E74A variant is catalytically inactive. The luciferase activity of reporter gene co-transfected with 6F6 was set to 100%. The error bars indicate the standard deviations of at least three independent experiments. (B) Competition between 6F6-3a and 6F6. To investigate the targeting function of the zinc-finger DBD, constant amounts of IE175-Luc and 6F6-3a were co-transfected with different concentrations of a construct expressing only 6F6, while the total amount of DNA in each transfection was normalized by adding an empty vector.
Figure 5
(A) Methylation status of HSV-1 DNA isolated during progressing viral infection. Analyses were performed at 30 h after infection of COS-7 cells transiently expressing targeted DNA Mtases (6F6-3a or B1-3a) or control proteins (6F6-KOX or catalytically inactive 6F6-3a-E74A). The analyzed region corresponds to IE175k promoter sequence containing the B1/6F6 binding site. Each line represents an individual clone. The empty squares represent unmethylated CG sites, whereas the filled ones represent methylated sites. (B) Comparison of the methylation levels of clones obtained in the bisulfite methylation analysis of the IE175k promoter region of HSV-1 DNA after infection of COS-7 cells expressing targeted DNA MTases [examples of the data are shown in (A)]. Methylation levels were corrected for the incomplete bisulfite conversion, which was between 2 and 10% in each case.
Figure 6
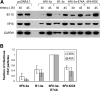
Inhibition of viral lytic cycle by zinc-finger MTase fusion constructs. (A) western blot analysis of the expression of HSV-1 antigens during the course of low multiplicity infection with HSV-1. Cells were transfected with zinc finger MTase or control constructs expressing 6F6-3a, B1-3a, 6F6-3a-E74A or 6F6-KOX (as indicted), infected with wt HSV-1 and harvested after 30 or 45 h p.i. Expression of HSV-1 VP16 was detected with mAb LP1 while expression of HSV-1 IE110k was detected using r191 antibody. Equal loading of the samples was verified using antibody against cellular protein GAPDH. (B) Inhibition of HSV-1 propagation by 6F6-3a and B1-3a. Culture medium samples harvested at 30 and 45 h after the infection with HSV-1 at low m.o.i were used for plaque assays on confluent monolayers of COS-7 cells, in 10-fold serial dilutions of the virus. The graph shows relative number of infectious HSV-1 particles released into the medium at indicated times p.i. from cells expressing active MTases (6F6-3a and B1-3a), catalytically inactive 6F6-3a-E74A mutant or 6F6-KOX fusion proteins. The error bars give the standard deviation of at least three independent experiments.
Similar articles
- Targeted methylation and gene silencing of VEGF-A in human cells by using a designed Dnmt3a-Dnmt3L single-chain fusion protein with increased DNA methylation activity.
Siddique AN, Nunna S, Rajavelu A, Zhang Y, Jurkowska RZ, Reinhardt R, Rots MG, Ragozin S, Jurkowski TP, Jeltsch A. Siddique AN, et al. J Mol Biol. 2013 Feb 8;425(3):479-91. doi: 10.1016/j.jmb.2012.11.038. Epub 2012 Dec 4. J Mol Biol. 2013. PMID: 23220192 - DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters.
Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. Robertson KD, et al. Nat Genet. 2000 Jul;25(3):338-42. doi: 10.1038/77124. Nat Genet. 2000. PMID: 10888886 - Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor.
Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, Fuks F, Lo Coco F, Kouzarides T, Nervi C, Minucci S, Pelicci PG. Di Croce L, et al. Science. 2002 Feb 8;295(5557):1079-82. doi: 10.1126/science.1065173. Science. 2002. PMID: 11834837 - Structure, function and mechanism of exocyclic DNA methyltransferases.
Bheemanaik S, Reddy YV, Rao DN. Bheemanaik S, et al. Biochem J. 2006 Oct 15;399(2):177-90. doi: 10.1042/BJ20060854. Biochem J. 2006. PMID: 16987108 Free PMC article. Review. - Expression of exogenous DNA methyltransferases: application in molecular and cell biology.
Dyachenko OV, Tarlachkov SV, Marinitch DV, Shevchuk TV, Buryanov YI. Dyachenko OV, et al. Biochemistry (Mosc). 2014 Feb;79(2):77-87. doi: 10.1134/S0006297914020011. Biochemistry (Mosc). 2014. PMID: 24794723 Review.
Cited by
- Synthetic epigenetics-towards intelligent control of epigenetic states and cell identity.
Jurkowski TP, Ravichandran M, Stepper P. Jurkowski TP, et al. Clin Epigenetics. 2015 Mar 4;7(1):18. doi: 10.1186/s13148-015-0044-x. eCollection 2015. Clin Epigenetics. 2015. PMID: 25741388 Free PMC article. - Reshaping the Landscape of the Genome: Toolkits for Precise DNA Methylation Manipulation and Beyond.
Zhu C, Hao Z, Liu D. Zhu C, et al. JACS Au. 2023 Dec 21;4(1):40-57. doi: 10.1021/jacsau.3c00671. eCollection 2024 Jan 22. JACS Au. 2023. PMID: 38274248 Free PMC article. Review. - Specific zinc finger-induced methylation of PD-L1 promoter inhibits its expression.
Li X, Wang Z, Huang J, Luo H, Zhu S, Yi H, Zheng L, Hu B, Yu L, Li L, Xie J, Zhu N. Li X, et al. FEBS Open Bio. 2019 Jun;9(6):1063-1070. doi: 10.1002/2211-5463.12568. Epub 2019 May 14. FEBS Open Bio. 2019. PMID: 31090214 Free PMC article. - Large-scale manipulation of promoter DNA methylation reveals context-specific transcriptional responses and stability.
de Mendoza A, Nguyen TV, Ford E, Poppe D, Buckberry S, Pflueger J, Grimmer MR, Stolzenburg S, Bogdanovic O, Oshlack A, Farnham PJ, Blancafort P, Lister R. de Mendoza A, et al. Genome Biol. 2022 Jul 26;23(1):163. doi: 10.1186/s13059-022-02728-5. Genome Biol. 2022. PMID: 35883107 Free PMC article. - Transcriptional silencing and reactivation in transgenic zebrafish.
Goll MG, Anderson R, Stainier DY, Spradling AC, Halpern ME. Goll MG, et al. Genetics. 2009 Jul;182(3):747-55. doi: 10.1534/genetics.109.102079. Epub 2009 May 11. Genetics. 2009. PMID: 19433629 Free PMC article.
References
- Reik W., Dean W., Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. - PubMed
- Jones P.A., Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. - PubMed
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
- Felsenfeld G., Groudine M. Controlling the double helix. Nature. 2003;421:448–453. - PubMed
- Chen W.G., Chang Q., Lin Y., Meissner A., West A.E., Griffith E.C., Jaenisch R., Greenberg M.E. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases