How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria - PubMed (original) (raw)
Review
How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria
Josef Deutscher et al. Microbiol Mol Biol Rev. 2006 Dec.
Erratum in
- Microbiol Mol Biol Rev. 2008 Sep;72(3):555
Abstract
The phosphoenolpyruvate(PEP):carbohydrate phosphotransferase system (PTS) is found only in bacteria, where it catalyzes the transport and phosphorylation of numerous monosaccharides, disaccharides, amino sugars, polyols, and other sugar derivatives. To carry out its catalytic function in sugar transport and phosphorylation, the PTS uses PEP as an energy source and phosphoryl donor. The phosphoryl group of PEP is usually transferred via four distinct proteins (domains) to the transported sugar bound to the respective membrane component(s) (EIIC and EIID) of the PTS. The organization of the PTS as a four-step phosphoryl transfer system, in which all P derivatives exhibit similar energy (phosphorylation occurs at histidyl or cysteyl residues), is surprising, as a single protein (or domain) coupling energy transfer and sugar phosphorylation would be sufficient for PTS function. A possible explanation for the complexity of the PTS was provided by the discovery that the PTS also carries out numerous regulatory functions. Depending on their phosphorylation state, the four proteins (domains) forming the PTS phosphorylation cascade (EI, HPr, EIIA, and EIIB) can phosphorylate or interact with numerous non-PTS proteins and thereby regulate their activity. In addition, in certain bacteria, one of the PTS components (HPr) is phosphorylated by ATP at a seryl residue, which increases the complexity of PTS-mediated regulation. In this review, we try to summarize the known protein phosphorylation-related regulatory functions of the PTS. As we shall see, the PTS regulation network not only controls carbohydrate uptake and metabolism but also interferes with the utilization of nitrogen and phosphorus and the virulence of certain pathogens.
Figures
FIG. 1.
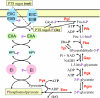
Carbohydrate transport and phosphorylation by the PTS and their coupling to glycolysis. Carbohydrates are transported and concomitantly phosphorylated by the PTS. The phosphorylated carbohydrate feeds into glycolysis, normally at the glucose-6-P or fructose-6-P level. Two phosphoenolpyruvate molecules are usually formed in glycolysis, one of which is used to drive the transport and initial phosphorylation of the carbohydrate. As a result, the phosphorylation state of the PTS proteins depends on both the concentration of extracellular carbohydrates and the ratio of internal phosphoenolpyruvate and pyruvate. Abbreviations for enzymes (in boldface type) are as follows: Pgi, phosphoglucose isomerase; Pfk, phosphofructokinase; Fba, fructose-1,6-bisphosphate aldolase; Tpi, triose-phosphate isomerase; Gap, glyceraldehyde-3-phosphate dehydrogenase; Pgk, phosphoglycerate kinase; Pgm, phosphoglycerate mutase; Eno, enolase; Pyk, pyruvate kinase.
FIG. 2.
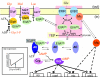
Mechanisms underlying CCR and inducer exclusion in enteric bacteria. The import of glucose and other PTS carbohydrates leads to net dephosphorylation of the PTS proteins (including EIIAGlc and the B domain of EIIBCGlc) and thereby to inducer exclusion and recruitment of the transcription regulator Mlc to the membrane. The upper left part of the figure shows that unphosphorylated EIIAGlc blocks the import of lactose, maltose, and melibiose and the phosphorylation of glycerol by binding to the respective transporter or kinase. The upper right part of the figure shows recruitment of Mlc by unphosphorylated EIIBCGlc, which prevents the regulator from binding to its target sites on the DNA. In the absence of glucose and in the presence of phosphoenolpyruvate, the PTS proteins are found mainly in the phosphorylated state. The central part of the figure shows that phosphorylated EIIAGlc activates adenylate cyclase (AC) but probably only in the presence of an unknown adenylate cyclase activation factor (ACAF). Adenylate cyclase binds phosphorylated as well as unphosphorylated EIIAGlc (see reference 632). The bottom part of the figure shows the effect of the activated transcription factors (free Mlc and Crp:cAMP) on the transcription of the genes encoding Crp, adenylate cyclase, Mlc, and EIIBCGlc, respectively. The inset shows the Crp:cAMP concentration dependence of crp transcription (deduced from data reported in references and 311). The arrow indicates the physiological concentration of activated Crp in exponentially growing cells in the absence of PTS carbohydrates.
FIG. 3.
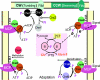
Mechanism of chemotaxis in E. coli and role of the PTS in carbohydrate chemotaxis. In the absence of chemotactically active molecules (top left), CheA autophosphorylates at a histidyl residue. This reaction is stimulated by CheW, which recruits CheA to the MCP receptor protein. P∼CheA transfers its phosphoryl group to CheY, and P∼CheY binds the flagellar motor FliM, thereby evoking clockwise (CW) rotation of the flagella, which leads to tumbling of the bacterium. In the presence of chemotactically active molecules (top right), the autophosphorylation activity of CheA is inhibited, and as a result, the concentration of P∼CheY drops. FliM molecules will no longer be complexed with CheY, which favors counterclockwise (CCW) rotation of the flagella. This results in smooth swimming towards increasing concentrations of the chemotactically active substance. The presence of an efficiently metabolizable PTS carbohydrate (or the absence of PEP) induces a similar response, as under these conditions, EI is present mainly in an unphosphorylated form. In fact, dephospho-EI inhibits the autophosphorylation of CheA (center) and therefore also favors smooth swimming. The sensitivity of the system towards the signal is regulated through methylation and demethylation of the MCPs (bottom), which are catalyzed by CheR and CheB, respectively. MCP methylation stimulates the autophosphorylation of CheA, and demethylation inhibits it.
FIG. 4.
Alignment of the first 55 amino acids of HPr proteins from firmicutes (B.s., B. subtilis; E.f., E. faecalis; L.c., L. casei; S.s., S. salivarius; S.c., S. carnosus), from a spirochete (T.p., T. pallidum), from proteobacteria with a Ser-46 region strongly resembling the corresponding sequence in HPr of firmicutes (X.f., Xylella fastidiosa; N.m., N. meningitidis), and from other gram-negative bacteria (E.c., E. coli; H.i., H. influenzae; V.c., V. cholerae) (V. cholerae possesses a second HPr in which the region around Ser-46 more strongly resembles the corresponding region in the HPr of firmicutes). The arrows indicate the amino acids His-15 and Ser-46. In HPrs from organisms, in which these residues are phosphorylated, they are shown in boldface type. Conserved regions around the phosphorylation sites are boxed.
FIG. 5.
The gene context of hprK in bacteria of the phylum Firmicutes. The hprK gene is followed by lgt in all sequenced genomes of the firmicutes except in L. mesenteroides, Oenococcus oeni, and some clostridiae. In addition, there are other genes associated with hprK that appear to be conserved. They include two genes without a known function (in L. lactis, E. faecalis, and several streptococci [S. agalactiae, S. mutans, S. pneumoniae, S. pyogenes, S. suis, S. thermophilus, and _S. uberis_]) and a glycerol-3-phosphate dehydrogenase gene and a UTP-glucose-1-phosphate uridylyltransferase gene (in E. faecalis) followed by a thioredoxin reductase gene (in L. acidophilus, L. johnsonii, L. gasseri [these species lack the uridylyltransferase], L. brevis, L. plantarum, P. pentosaceus, L. mesenteroides, and O. oeni [the latter two species lack the lgt gene]). Staphylococci contain YvoF, a protein with a hexapeptide transferase motif (S. aureus, S. epidermidis, S. haemolyticus, and S. saprophyticus). YvoF, together with YvoE (a pyrophosphatase), is also present in B. anthracis, B. cereus, B. thuringiensis, Exiguobacterium species, L. innocua, and L. monocytogenes, while YvoD, YvoE, and YvoF are found in the bacilli B. clausii, B. halodurans, B. licheniformis, B. stearothermophilus, B. subtilis, G. kaustophilus, and O. iheyensis.
FIG. 6.
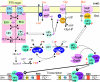
Mechanisms underlying CCR in firmicutes. The uptake of glucose and other rapidly metabolizable PTS sugars (top left) leads to a net dephosphorylation of the PTS proteins. The center and bottom of the figure show that the high concentration of FBP present in cells growing on a rapidly metabolizable carbohydrate stimulates the HPr kinase activity of the bifunctional HprK/P and the formation of P-Ser-HPr. P-Ser-HPr interacts with CcpA, and the protein complex binds to the cre operator sites on the DNA. The promoter regions are indicated as −10 and −35. CCA occurs when the cre is located upstream from the promoter, while CCR requires a cre located within or downstream from the promoter. High concentrations of Pi present in resting cells favor the pyrophosphate-producing dephosphorylation of P-Ser-HPr by HprK/P. The top right part of the figure shows that P-Ser-HPr probably interacts with certain non-PTS carbohydrate transport systems, such as the maltose transporter from L. casei, and thereby inhibits their transport activity. Phosphorylation of LacS by P∼His-HPr stimulates the lactose/galactose exchange reaction (in S. thermophilus). In the presence of rapidly metabolizable PTS substrates, the low level of P∼His-HPr does not allow sufficient phosphorylation of GlpK, which leads to the inactivation of GlpK and to inducer exclusion. Pyr., pyruvate.
FIG. 7.
Domain structure of the transcription antiterminator LicT and the transcription activators LicR and LevR of B. subtilis. The N-terminal RNA or DNA binding domain and the two regulatory domains PRD1 and PRD2 together with their conserved histidyl residues are indicated. Histidyl residues, which have been shown to become phosphorylated, are in boldface type. When the phosphorylation exerts a positive effect on the activity of the regulator, they are shown in red, whereas blue indicates the sites of negative regulation. For LicT, its four conserved histidyl residues become phosphorylated. Phosphorylation in PRD1 inhibits, while phosphorylation in PRD2 stimulates, LicT activity. Most other antiterminators of the BglG/SacY family also contain four conserved histidines, but sometimes, not all of them can be phosphorylated. LevR contains an NtrC-like central domain and EIIAMan- and EIIBGat-like domains inserted between the complete PRD1 and a truncated PRD2. The positive site of phosphorylation is His-585, and the negative site is His-869. The conserved histidyl residues His-506 and His-567 in PRD1 and the EIIBGat phosphorylation site (Cys-718) do not seem to become phosphorylated. An identical organization can be observed in most other NifA/NtrC-type PRD-containing transcription activators. However, a few LevR-like regulators contain a complete PRD2 with an additional potentially phosphorylatable histidyl residue. The DNA binding domain of LicR resembles that of DeoR, and PRD1 and PRD2 are followed by an EIIBGat-like and EIIAMtl-like domain. An identical domain organization can be observed in other DeoR-type PRD-containing transcription activators. All four conserved histidines in PRD1 and PRD2 are sites of positive regulation, while LicR activity is inhibited by phosphorylation at His-559 in the EIIAMtl-like domain.
FIG. 8.
Transcription regulation by the PTS via PRD-containing transcription antiterminators. (A) In the absence of the corresponding inducer, full-length transcription of several PTS-encoding genes/operons is inhibited owing to the formation of a terminator structure (t, yellow) on the nascent mRNA upstream from the start codon. Under these conditions, the corresponding antiterminator cannot bind to its RNA target, RAT (blue), because the EIIB is mainly phosphorylated and transfers its phosphoryl group to PRD1 of the antiterminator. The absence of a repressing sugar is expected to also allow phosphorylation at the activating domain (PRD2) by P∼His-HPr. However, the negative effect of phosphorylation at PRD1 is dominant. The RAT sequence can also form a stem-loop, which, however, was calculated to free less energy than the terminator t. Interestingly, in most antiterminator-controlled PTS operons, the two sites RAT and t overlap (green), and the formation of the terminator therefore prevents the formation of the RAT stem-loop and vice versa. (B) If an inducer is present, the EIIB as well as PRD1 of the corresponding antiterminator will be present mainly in an unphosphorylated form. Because antiterminator-controlled PTSs are usually low-capacity sugar transporters, there will be sufficient P∼His-HPr to guarantee activating phosphorylation in PRD2. The activated antiterminator binds to its RAT and thus favors the formation of the RAT stem-loop, thereby preventing the formation of the terminator stem-loop, as part of it (in green) is already used for the RAT stem-loop. (C) If, in addition to the inducing sugar, a repressing carbohydrate is present, the amount of P∼His-HPr will be low in the cells. In firmicutes, P-Ser-HPr will also be formed, which further lowers the amount of P∼His-HPr. These conditions prevent activating phosphorylation at PRD2, and most antiterminators are therefore inactive, although the presence of the inducer probably prevents the phosphorylation in PRD1. Dephosphorylation of PRD2 in the presence of a rapidly metabolizable PTS sugar therefore represents a CcpA-independent, P-Ser-HPr-dependent CCR mechanism. ptsH1 as well as licT(Pia) but not ccpA mutants are relieved from this type of CCR.
FIG. 9.
The Dha PTS of E. coli. Phosphotransfer from PEP to Dha is mediated by five distinct proteins (EI, HPr, DhaM, DhaL, and DhaK). DhaM itself is composed of three PTS domains: a truncated EI, HPr, and an EIIA of the mannose class PTS. From the P∼EIIA domain of DhaM, the phosphoryl group is transferred to an ADP molecule tightly bound to DhaL and from there is transferred to a Dha molecule bound to DhaK. Expression of the E. coli dha operon is regulated by the transcription activator DhaR. In addition, the gene encoding the activator DhaR is subject to negative autoregulation. When Dha is present in the cell, it is rapidly phosphorylated, and the ultimate phosphoryl donor, DhaL, therefore carries predominantly ADP. The DhaL:ADP complex binds to DhaR and stimulates its regulator functions. DhaK without Dha interacts with DhaR and down-regulates its activity, while the complex with its substrate, which is formed when Dha is present in the cell, does not interact with DhaR (37). It should be noted that many other organisms do not contain the three-domain DhaM but possess only EIIADha instead. In these bacteria, HPr probably transfers its phosphoryl group directly to EIIADha.
FIG. 10.
Gene organization downstream from rpoN in several bacteria. The genes encode the sigma factor σ54 (rpoN), the ribosome-associated protein Y (yhbH, in yellow), EIIANtr (ptsN), an ATP-binding protein (yhbJ, in red), and the HPr paralog NPr (npr), respectively.
References
- Ab, E., G. Schuurman-Wolters, J. Reizer, M. H. Saier, Jr., K. Dijkstra, R. M. Scheek, and G. T. Robillard. 1997. The NMR side-chain assignments and solution structure of enzyme IIB(cellobiose) of the phosphoenolpyruvate-dependent phosphotransferase system of Escherichia coli. Protein Sci. 6:304-314. - PMC - PubMed
- Ab, E., G. K. Schuurman-Wolters, D. Nijlant, K. Dijkstra, M. H. Saier, Jr., G. T. Robillard, and R. M. Scheek. 2001. NMR structure of cysteinyl-phosphorylated enzyme IIB of the N,N′-diacetylchitobiose-specific phosphoenolpyruvate-dependent phosphotransferase system of Escherichia coli. J. Mol. Biol. 308:993-1009. - PubMed
- Abramson, J., I. Smirnova, V. Kasho, G. Verner, S. Iwata, and H. R. Kaback. 2003. The lactose permease of Escherichia coli: overall structure, the sugar-binding site and the alternating access model for transport. FEBS Lett. 555:96-101. - PubMed
- Abramson, J., I. Smirnova, V. Kasho, G. Verner, H. R. Kaback, and S. Iwata. 2003. Structure and mechanism of the lactose permease of Escherichia coli. Science 301:610-615. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous