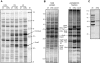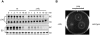The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium - PubMed (original) (raw)
The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium
Alexandra Sittka et al. Mol Microbiol. 2007 Jan.
Abstract
The RNA chaperone, Hfq, plays a diverse role in bacterial physiology beyond its original role as a host factor required for replication of Qbeta RNA bacteriophage. In this study, we show that Hfq is involved in the expression and secretion of virulence factors in the facultative intracellular pathogen, Salmonella typhimurium. A Salmonella hfq deletion strain is highly attenuated in mice after both oral and intraperitoneal infection, and shows a severe defect in invasion of epithelial cells and a growth defect in both epithelial cells and macrophages in vitro. Surprisingly, we find that these phenotypes are largely independent of the previously reported requirement of Hfq for expression of the stationary phase sigma factor, RpoS. Our results implicate Hfq as a key regulator of multiple aspects of virulence including regulation of motility and outer membrane protein (OmpD) expression in addition to invasion and intracellular growth. These pleiotropic effects are suggested to involve a network of regulatory small non-coding RNAs, placing Hfq at the centre of post-transcriptional regulation of virulence gene expression in Salmonella. In addition, the hfq mutation appears to cause a chronic activation of the RpoE-mediated envelope stress response which is likely due to a misregulation of membrane protein expression.
Figures
Fig. 1
Details of Salmonella hfq mutants and their growth characteristics. A. Genomic location of hfq in SL1344. The region cloned on complementation plasmid, pStHfq-6H, is indicated. B. Schematic representation of the insertion sites of the cat resistance cassette in the deletion mutant Δ_hfq_, the control strain hfq_-C, and the chromosomally HIS-tagged strain, hfq_HIS. C and D. Growth and cell viability of hfq mutant strains (open squares: wild-type; filled triangles: hfq_HIS; open diamonds: hfq_-C; stars: Δ_hfq). (C) OD600 values of triplicate cultures in LB medium were determined in 45 min intervals. (D) Bacteria were plated to determine viable counts (from triplicate cultures) at an OD of 0.3 and of 2, and 6 h after cultures had reached an OD of 2. E. Complementation of the slight growth defect of the Δ_hfq strain by plasmid pStHfq-6H (open squares: wild-type strain carrying control plasmid pVP012; stars: Δ_hfq carrying a control plasmid; filled circles: Δ_hfq complemented with pStHfq-6H).
Fig. 2
The Δhfq mutant is severely attenuated in mice. A. Groups of five Balb/c mice were infected perorally with suspensions of ∼108 bacteria of either the wild-type or Δ_hfq_ strains. Bacterial loads in spleen homogenates were determined 72 h post infection. For intraperitoneal infections (B) 1:1 mixtures of both, wild-type and Δ_hfq_ strain, each strain at ∼105 bacteria, were used for infections. Forty-eight hours post infection, spleens were removed and the cfu ml−1 for each strain was determined in spleen homogenates by plating to selective plates for calculation of the relative ratios of the two, co-infecting strains (competitive index, CI, see text).
Fig. 3
Altered protein expression in Salmonella Δhfq. SDS-PAGE (10–12% gels) of protein samples of SL1344 wild-type and Δ_hfq_ prepared from different growth phases (LOG: logarithmic phase, OD600 of 0.3; ES: early stationary phase, OD600 of 2; LS: late stationary phase, 6 h after cells had reached an OD600 of 2). A. Total protein samples. B. Total protein and periplasmic fractions; samples of a Δ_rpoS_ strain were included as an additional control. C. Secreted protein fractions of early stationary phase bacteria.
Fig. 5
SPI1-inducing conditions restore effector levels and their secretion in the Δ_hfq_ strain. A. Comparison of secreted proteins of wild-type, Δ_hfq_, Δ_spi1_ and Δ_rpoS_ grown for 12 h under standard conditions (lanes 1–4) or SPI1-inducing conditions (lanes 5–8) by SDS-PAGE analysis. B. Western blot detection of effector and needle proteins in total protein samples and secreted fractions of bacteria grown for 12 h under SPI1-inducing conditions. Bacterial strains from left to right: wild-type, Δ_hfq_, Δ_spi1_.
Fig. 4
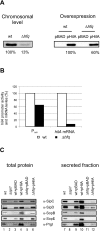
The hfq deletion mutant is impaired in HilA expression and shows reduced effector levels. A. HilA levels in wild-type and Δ_hfq Salmonella_ grown to early stationary phase. Shown are Western blots probed for chromosomally encoded HilAFLAG protein (left panel), or HilAmyc protein as expressed from pBAD-HilA expression plasmid (right panel). Bacteria carrying the empty pBAD vector were included as control. B. hilA promoter activity determined with a transcriptional hilA-gfp fusion in early stationary phase (P_hilA_), and hilA mRNA levels as determined by Northern analysis. Given are relative values obtained for Δ_hfq_, with the levels determined for the wild-type strain set to 100%. C. Western blot detection of effector and needle proteins in total protein samples and secreted fractions of bacteria grown to early stationary phase. Bacterial strains from left to right: wild-type, Δ_spi1,_ wild-type strain carrying a pBAD control vector, wild-type strain carrying a pBAD-HilA expression plasmid, Δ_hfq_ carrying a pBAD control vector, Δ_hfq_ with pBAD-HilA expression plasmid. All strains were grown in LB medium complemented with 0.05%
l
-arabinose to facilitate HilA expression from plasmid pBAD-HilA.
Fig. 6
The Δ_hfq_ strain is non-motile. A. Northern blot detection of fliC mRNA levels in wild-type and Δ_hfq_ cells at logarithmic and early stationary phase before and within 32 min after rifampicin treatment. Densitometry of the Northern blot signals showed that the fliC mRNA decays with the same half-life in both genetic backgrounds (∼9 min or ∼7 min in logarithmic or early stationary phase cultures respectively). 5S signals are shown as loading control. B. To measure motility, equal numbers of bacteria from each strain were inoculated onto a motility agar-plate. The image was obtained following 4 h of incubation at 37°C.
Fig. 7
Hfq is essential for growth rate-dependent repression of OmpD. A. SDS-PAGE analysis of total protein prepared from wild-type, Δ_hfq_, Δ_ompD_ and Δ_hfq_/Δ_ompD_ bacteria grown to early stationary phase. OmpD protein levels as quantified by fluorescent staining (not shown) are given below each lane. B. Northern blot detection of ompC and ompD mRNA levels of wild-type and Δ_hfq_ bacteria grown to either logarithmic or early stationary phase prior to (0 min) and within 32 min of rifampicin treatment. 5S sRNA probing (loading control) is shown below each panel. C. Decay of ompC and ompD mRNA upon rifampicin treatment as derived from quantification of the Northern blot signals shown in (B). Logarithmic phase, wild-type (filled circles) or Δ_hfq_ (open circles); early stationary phase, wild-type (filled squares) or Δ_hfq_ (open squares). D. Hfq binds to ompD 5′ UTR RNA in vitro (gel mobility shift assay). Left panel: 1 nM of 32P-labelled ompD was incubated with increasing concentrations of Hfq protein (given above the lanes). Following a 15 min incubation at 37°C samples were run on a native 6% gel. Shown is an autoradiograph of the gel. A control gel shift assay with an Hfq-independent RNA derived from the metK 5′ UTR is shown in the right panel.
Fig. 8
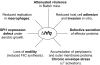
Summary of phenotypes of the Salmonella hfq mutation determined in this study.
Similar articles
- Loss of Hfq activates the sigmaE-dependent envelope stress response in Salmonella enterica.
Figueroa-Bossi N, Lemire S, Maloriol D, Balbontín R, Casadesús J, Bossi L. Figueroa-Bossi N, et al. Mol Microbiol. 2006 Nov;62(3):838-52. doi: 10.1111/j.1365-2958.2006.05413.x. Epub 2006 Sep 25. Mol Microbiol. 2006. PMID: 16999834 - Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression.
Ding Y, Davis BM, Waldor MK. Ding Y, et al. Mol Microbiol. 2004 Jul;53(1):345-54. doi: 10.1111/j.1365-2958.2004.04142.x. Mol Microbiol. 2004. PMID: 15225327 - Global systems-level analysis of Hfq and SmpB deletion mutants in Salmonella: implications for virulence and global protein translation.
Ansong C, Yoon H, Porwollik S, Mottaz-Brewer H, Petritis BO, Jaitly N, Adkins JN, McClelland M, Heffron F, Smith RD. Ansong C, et al. PLoS One. 2009;4(3):e4809. doi: 10.1371/journal.pone.0004809. Epub 2009 Mar 11. PLoS One. 2009. PMID: 19277208 Free PMC article. - [The regulatory role of the Hfq protein in bacterial cells].
Vasil'eva IuM, Garber MB. Vasil'eva IuM, et al. Mol Biol (Mosk). 2002 Nov-Dec;36(6):970-7. Mol Biol (Mosk). 2002. PMID: 12500533 Review. Russian. - Hfq: a multifaceted RNA chaperone involved in virulence.
Feliciano JR, Grilo AM, Guerreiro SI, Sousa SA, Leitão JH. Feliciano JR, et al. Future Microbiol. 2016;11(1):137-51. doi: 10.2217/fmb.15.128. Epub 2015 Dec 18. Future Microbiol. 2016. PMID: 26685037 Review.
Cited by
- Metabolic Proteins Expression Up-Regulated in Blood-Borne Extensively Drug-Resistant Salmonella Typhi Isolates from Pakistan.
Yasin N, Rahman H, Qasim M, Nisa I, Sarwar Y, Khan N, Alzahrani KJ, Alsuwat MA, Alzahrani FM, Aljohani A. Yasin N, et al. Medicina (Kaunas). 2024 Aug 27;60(9):1404. doi: 10.3390/medicina60091404. Medicina (Kaunas). 2024. PMID: 39336445 Free PMC article. - Microbiology of human spaceflight: microbial responses to mechanical forces that impact health and habitat sustainability.
Nickerson CA, McLean RJC, Barrila J, Yang J, Thornhill SG, Banken LL, Porterfield DM, Poste G, Pellis NR, Ott CM. Nickerson CA, et al. Microbiol Mol Biol Rev. 2024 Sep 26;88(3):e0014423. doi: 10.1128/mmbr.00144-23. Epub 2024 Aug 19. Microbiol Mol Biol Rev. 2024. PMID: 39158275 Review. - RNA interactome of hypervirulent Klebsiella pneumoniae reveals a small RNA inhibitor of capsular mucoviscosity and virulence.
Wu K, Lin X, Lu Y, Dong R, Jiang H, Svensson SL, Zheng J, Shen N, Camilli A, Chao Y. Wu K, et al. Nat Commun. 2024 Aug 13;15(1):6946. doi: 10.1038/s41467-024-51213-z. Nat Commun. 2024. PMID: 39138169 Free PMC article. - The RNA chaperone Hfq has a multifaceted role in Edwardsiella ictaluri.
Akgul A, Kalindamar S, Kordon AO, Abdelhamed H, Ibrahim I, Tekedar HC, Karsi A. Akgul A, et al. Front Cell Infect Microbiol. 2024 Jul 19;14:1394008. doi: 10.3389/fcimb.2024.1394008. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39099884 Free PMC article. - Characterization of a Salmonella enterica serovar Typhimurium lineage with rough colony morphology and multidrug resistance.
Xiang Y, Zhu K, Min K, Zhang Y, Liu J, Liu K, Han Y, Li X, Du X, Wang X, Huang Y, Li X, Peng Y, Yang C, Liu H, Liu H, Li X, Wang H, Wang C, Wang Q, Jia H, Yang M, Wang L, Wu Y, Cui Y, Chen F, Yang H, Baker S, Xu X, Yang J, Song H, Qiu S. Xiang Y, et al. Nat Commun. 2024 Jul 20;15(1):6123. doi: 10.1038/s41467-024-50331-y. Nat Commun. 2024. PMID: 39033143 Free PMC article.
References
- Ahmer BM, van Reeuwijk J, Watson PR, Wallis TS, Heffron F. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol Microbiol. 1999;31:971–982. - PubMed
- van Asten FJ, Hendriks HG, Koninkx JF, van Dijk JE. Flagella-mediated bacterial motility accelerates but is not required for Salmonella serotype Enteritidis invasion of differentiated Caco-2 cells. Int J Med Microbiol. 2004;294:395–399. - PubMed
- Bajaj V, Lucas RL, Hwang C, Lee CA. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. - PubMed
- Bang IS, Frye JG, McClelland M, Velayudhan J, Fang FC. Alternative sigma factor interactions in Salmonella: sigma and sigma promote antioxidant defences by enhancing sigma levels. Mol Microbiol. 2005;56:811–823. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources