Solving structures of protein complexes by molecular replacement with Phaser - PubMed (original) (raw)
Solving structures of protein complexes by molecular replacement with Phaser
Airlie J McCoy. Acta Crystallogr D Biol Crystallogr. 2007 Jan.
Abstract
Molecular replacement (MR) generally becomes more difficult as the number of components in the asymmetric unit requiring separate MR models (i.e. the dimensionality of the search) increases. When the proportion of the total scattering contributed by each search component is small, the signal in the search for each component in isolation is weak or non-existent. Maximum-likelihood MR functions enable complex asymmetric units to be built up from individual components with a ;tree search with pruning' approach. This method, as implemented in the automated search procedure of the program Phaser, has been very successful in solving many previously intractable MR problems. However, there are a number of cases in which the automated search procedure of Phaser is suboptimal or encounters difficulties. These include cases where there are a large number of copies of the same component in the asymmetric unit or where the components of the asymmetric unit have greatly varying B factors. Two case studies are presented to illustrate how Phaser can be used to best advantage in the standard ;automated MR' mode and two case studies are used to show how to modify the automated search strategy for problematic cases.
Figures
Figure 1
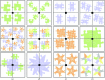
Catalogue of some possible contents of the unit cell for a crystal of space group _P_4. The contents of the asymmetric unit are as follows: top row, (a) one monomer, (b) two monomers, (c) biological homodimer, (d) two biological homodimers; middle row, (a) three biological heterodimers, (b) biological heterotetramer, (c) biological homotetramer, (d) one monomer of a biological homotetramer; bottom row, (a) one heterodimer of a biological hetero-octamer, (b) two monomers of a biological homo-octamer, (c) biological homopentamer, (d) biological heteropentamer.
Figure 2
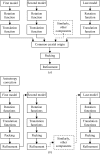
Flow diagrams for solving structures of protein complexes by MR. (a) Traditional MR, where each search component must be found separately and then combined to assemble the asymmetric unit. (b) Maximum-likelihood MR, where placement of the first component is used to aid the search for the second and subsequent components; the complete asymmetric unit is generated by the addition of search components one at a time.
Figure 3
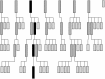
Tree search with pruning MR search strategy for a crystal with four search components in the asymmetric unit. Row 1 represents the results of the search for the first component, where seven of eight solutions meet the selection criteria. Row 2 represents the results from the search for the second component. The search is performed using the seven possible placements for the first component as the background for seven separate searches for the second component. 13 of the 22 results of the seven searches that do not meet the selection criteria are pruned from the search tree. At the end of this step, two of the four components have been placed in nine potential solutions. Row 3 represents the results from the search for the third component. As the percentage of the total scattering being modelled increases so does the signal-to-noise ratio of the search and there is better discrimination of the best solution in this step, where 17 of 23 branches are pruned. Row 4 represents the results of searching for the fourth and final component. The correct solution, which includes placements for all four components, stands out well above the noise. The history of this solution can be traced through the search tree (shown in black)
Figure 4
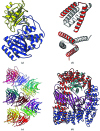
(a) Structure of the β-lactamase (BETA)–β-lactamase inhibitor (BLIP) complex. BETA is in blue and BLIP is in yellow. (b) Structure of the ROP four-helix bundle structure. The asymmetric unit is shown in red and crystallographically related molecules are shown in white. Together, they form two four-helix bundles. The search model was a 26-residue polyalanine helix. (c) The 15 molecules in the asymmetric unit for the Vκ antibody fibre. The molecules form a continuous fibre along the 64 axis in the crystals (space group _P_6422). (d) The AP2 complex of four proteins. The α subunit (a superhelix of α-helices) is shown in red, the β2 subunit in blue (a similar superhelix of helices), the σ2 subunit in cyan (mixed α-helix/β-sheet structure) and the μ2 subunit in magenta (which consists of an N-terminal domain structurally homologous to the σ2 subunit and a larger C-terminal mixed α-helix/β-sheet structure).
Similar articles
- Coping with strong translational noncrystallographic symmetry and extreme anisotropy in molecular replacement with Phaser: human Rab27a.
Jamshidiha M, Pérez-Dorado I, Murray JW, Tate EW, Cota E, Read RJ. Jamshidiha M, et al. Acta Crystallogr D Struct Biol. 2019 Mar 1;75(Pt 3):342-353. doi: 10.1107/S2059798318017825. Epub 2019 Feb 28. Acta Crystallogr D Struct Biol. 2019. PMID: 30950405 Free PMC article. - Improved estimates of coordinate error for molecular replacement.
Oeffner RD, Bunkóczi G, McCoy AJ, Read RJ. Oeffner RD, et al. Acta Crystallogr D Biol Crystallogr. 2013 Nov;69(Pt 11):2209-15. doi: 10.1107/S0907444913023512. Epub 2013 Oct 12. Acta Crystallogr D Biol Crystallogr. 2013. PMID: 24189232 Free PMC article. Clinical Trial. - Using Phaser and ensembles to improve the performance of SIMBAD.
Simpkin AJ, Simkovic F, Thomas JMH, Savko M, Lebedev A, Uski V, Ballard CC, Wojdyr M, Shepard W, Rigden DJ, Keegan RM. Simpkin AJ, et al. Acta Crystallogr D Struct Biol. 2020 Jan 1;76(Pt 1):1-8. doi: 10.1107/S2059798319015031. Epub 2020 Jan 1. Acta Crystallogr D Struct Biol. 2020. PMID: 31909738 Free PMC article. - Acknowledging Errors: Advanced Molecular Replacement with Phaser.
McCoy AJ. McCoy AJ. Methods Mol Biol. 2017;1607:421-453. doi: 10.1007/978-1-4939-7000-1_18. Methods Mol Biol. 2017. PMID: 28573584 Review. - Phaser.MRage: automated molecular replacement.
Bunkóczi G, Echols N, McCoy AJ, Oeffner RD, Adams PD, Read RJ. Bunkóczi G, et al. Acta Crystallogr D Biol Crystallogr. 2013 Nov;69(Pt 11):2276-86. doi: 10.1107/S0907444913022750. Epub 2013 Oct 18. Acta Crystallogr D Biol Crystallogr. 2013. PMID: 24189240 Free PMC article. Review.
Cited by
- Different 3D domain-swapped oligomeric cyanovirin-N structures suggest trapped folding intermediates.
Koharudin LM, Liu L, Gronenborn AM. Koharudin LM, et al. Proc Natl Acad Sci U S A. 2013 May 7;110(19):7702-7. doi: 10.1073/pnas.1300327110. Epub 2013 Apr 22. Proc Natl Acad Sci U S A. 2013. PMID: 23610431 Free PMC article. - Structural insight into the Scribble PDZ domains interaction with the oncogenic Human T-cell lymphotrophic virus-1 (HTLV-1) Tax1 PBM.
Javorsky A, Maddumage JC, Mackie ERR, Soares da Costa TP, Humbert PO, Kvansakul M. Javorsky A, et al. FEBS J. 2023 Feb;290(4):974-987. doi: 10.1111/febs.16607. Epub 2022 Sep 6. FEBS J. 2023. PMID: 36029163 Free PMC article. - Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite.
Cerutti G, Guo Y, Zhou T, Gorman J, Lee M, Rapp M, Reddem ER, Yu J, Bahna F, Bimela J, Huang Y, Katsamba PS, Liu L, Nair MS, Rawi R, Olia AS, Wang P, Zhang B, Chuang GY, Ho DD, Sheng Z, Kwong PD, Shapiro L. Cerutti G, et al. Cell Host Microbe. 2021 May 12;29(5):819-833.e7. doi: 10.1016/j.chom.2021.03.005. Epub 2021 Mar 12. Cell Host Microbe. 2021. PMID: 33789084 Free PMC article. - The structure of the Mycobacterium smegmatis trehalose synthase reveals an unusual active site configuration and acarbose-binding mode.
Caner S, Nguyen N, Aguda A, Zhang R, Pan YT, Withers SG, Brayer GD. Caner S, et al. Glycobiology. 2013 Sep;23(9):1075-83. doi: 10.1093/glycob/cwt044. Epub 2013 Jun 4. Glycobiology. 2013. PMID: 23735230 Free PMC article. - Structural characterization and computational analysis of PDZ domains in Monosiga brevicollis.
Gao M, Mackley IGP, Mesbahi-Vasey S, Bamonte HA, Struyvenberg SA, Landolt L, Pederson NJ, Williams LI, Bahl CD, Brooks L 3rd, Amacher JF. Gao M, et al. Protein Sci. 2020 Nov;29(11):2226-2244. doi: 10.1002/pro.3947. Epub 2020 Sep 25. Protein Sci. 2020. PMID: 32914530 Free PMC article.
References
- Collins, B. M., McCoy, A. J., Kent, H. M., Evans, P. & Owen, D. J. (2002). Cell, 109, 523–535. - PubMed
- Crowther, R. A. (1972). The Molecular Replacement Method, edited by M. G. Rossmann, pp. 173–178. New York: Gordon & Breach.
- Dauter, Z., Betzel, C., Genov, N., Pipon, N. & Wilson, K. S. (1991). Acta Cryst. B47, 707–730. - PubMed
- Fujinaga, M. & Read, R. J. (1987). J. Appl. Cryst. 20, 517–521.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources