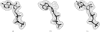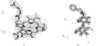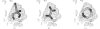Ligand identification using electron-density map correlations - PubMed (original) (raw)
Ligand identification using electron-density map correlations
Thomas C Terwilliger et al. Acta Crystallogr D Biol Crystallogr. 2007 Jan.
Abstract
A procedure for the identification of ligands bound in crystal structures of macromolecules is described. Two characteristics of the density corresponding to a ligand are used in the identification procedure. One is the correlation of the ligand density with each of a set of test ligands after optimization of the fit of that ligand to the density. The other is the correlation of a fingerprint of the density with the fingerprint of model density for each possible ligand. The fingerprints consist of an ordered list of correlations of each the test ligands with the density. The two characteristics are scored using a Z-score approach in which the correlations are normalized to the mean and standard deviation of correlations found for a variety of mismatched ligand-density pairs, so that the Z scores are related to the probability of observing a particular value of the correlation by chance. The procedure was tested with a set of 200 of the most commonly found ligands in the Protein Data Bank, collectively representing 57% of all ligands in the Protein Data Bank. Using a combination of these two characteristics of ligand density, ranked lists of ligand identifications were made for representative (F(o) - F(c))exp(i(phi)c) difference density from entries in the Protein Data Bank. In 48% of the 200 cases, the correct ligand was at the top of the ranked list of ligands. This approach may be useful in identification of unknown ligands in new macromolecular structures as well as in the identification of which ligands in a mixture have bound to a macromolecule.
Figures
Figure 1
(a) ATP fitted into model 2.5 Å density for ATP. (b) ddATP fitted into model density for ATP. (c) GTP fitted into model density for ATP.
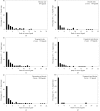
Figure 2
Histograms of rank position of correct ligands. (a) Scoring using correlation of density, considering 119 unique ligands. (b) Scoring using Z score derived from correlation of density. (c) Scoring using Z score derived from correlation of fingerprints of density and fingerprints of model density. (d) Scoring using sum of Z scores from correlation of density and correlation of fingerprints of density. (e) As in (d), but considering all 200 of the most common ligands in the PDB. (f) As in (d), but considering only 31 unique ligands.
Figure 3
(a) F o − F c difference density for bacteriochlorophyll a at 2.4 Å (PDB code
1ogv
; Katona et al., 2003 ▶), fitted with the same ligand from PDB entry
1dv6
(Axelrod et al., 2000 ▶). (b) Difference density for cyclohexyl-hexyl-β-
d
-maltoside at a resolution of 1.1 Å (PDB code
1ong
; Venkatesan et al., 2004 ▶), fitted with the same ligand from PDB entry
1q2p
(Nukaga et al., 2003 ▶).
Figure 4
Fitting of F o − F c difference density for tris-(hydroxylmethyl)-methane from PDB entry
1m6z
(A. Noergaard, P. Harris, S. Larsen & H. E. M. Christensen, unpublished) at a resolution of 1.4 Å. (a) Density fitted by the same ligand from a different PDB entry (
1s18
; Dai et al., 2004 ▶). (b) Density fitted with oxalate. (c) Density fitted with dioxane.
Figure 5
Fingerprints of difference density. (a) Correlation of each of 119 unique ligands after fitting to difference density for tris-(hydroxyamino)-methane from PDB entry
1m6z
(A. Noergaard, P. Harris, S. Larsen & H. E. M. Christensen, unpublished work) at a resolution of 1.4 Å. The ligands are sorted from left to right based on increasing numbers of non-H atoms. (b) As in (a), except fitting to difference density for ATP from PDB entry
1aq2
at a resolution of 1.9 Å (Tari et al., 1997 ▶). The correlations are all indicated by filled triangles, except for the correlation of the correct ligand, which is indicated by an open diamond.
Similar articles
- Assisted assignment of ligands corresponding to unknown electron density.
Binkowski TA, Cuff M, Nocek B, Chang C, Joachimiak A. Binkowski TA, et al. J Struct Funct Genomics. 2010 Mar;11(1):21-30. doi: 10.1007/s10969-010-9078-7. Epub 2010 Jan 21. J Struct Funct Genomics. 2010. PMID: 20091237 Free PMC article. - Validation of ligands in macromolecular structures determined by X-ray crystallography.
Smart OS, Horský V, Gore S, Svobodová Vařeková R, Bendová V, Kleywegt GJ, Velankar S. Smart OS, et al. Acta Crystallogr D Struct Biol. 2018 Mar 1;74(Pt 3):228-236. doi: 10.1107/S2059798318002541. Epub 2018 Mar 2. Acta Crystallogr D Struct Biol. 2018. PMID: 29533230 Free PMC article. - qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
van Zundert GCP, Hudson BM, de Oliveira SHP, Keedy DA, Fonseca R, Heliou A, Suresh P, Borrelli K, Day T, Fraser JS, van den Bedem H. van Zundert GCP, et al. J Med Chem. 2018 Dec 27;61(24):11183-11198. doi: 10.1021/acs.jmedchem.8b01292. Epub 2018 Dec 6. J Med Chem. 2018. PMID: 30457858 Free PMC article. - You are lost without a map: Navigating the sea of protein structures.
Lamb AL, Kappock TJ, Silvaggi NR. Lamb AL, et al. Biochim Biophys Acta. 2015 Apr;1854(4):258-68. doi: 10.1016/j.bbapap.2014.12.021. Epub 2014 Dec 29. Biochim Biophys Acta. 2015. PMID: 25554228 Free PMC article. Review. - Keep it together: restraints in crystallographic refinement of macromolecule-ligand complexes.
Steiner RA, Tucker JA. Steiner RA, et al. Acta Crystallogr D Struct Biol. 2017 Feb 1;73(Pt 2):93-102. doi: 10.1107/S2059798316017964. Epub 2017 Feb 1. Acta Crystallogr D Struct Biol. 2017. PMID: 28177305 Free PMC article. Review.
Cited by
- A Structural Comparison of Oral SARS-CoV-2 Drug Candidate Ibuzatrelvir Complexed with the Main Protease (Mpro) of SARS-CoV-2 and MERS-CoV.
Chen P, Van Oers TJ, Arutyunova E, Fischer C, Wang C, Lamer T, van Belkum MJ, Young HS, Vederas JC, Lemieux MJ. Chen P, et al. JACS Au. 2024 Jul 30;4(8):3217-3227. doi: 10.1021/jacsau.4c00508. eCollection 2024 Aug 26. JACS Au. 2024. PMID: 39211604 Free PMC article. - pH-dependent reaction triggering in PmHMGR crystals for time-resolved crystallography.
Purohit V, Steussy CN, Rosales AR, Critchelow CJ, Schmidt T, Helquist P, Wiest O, Mesecar A, Cohen AE, Stauffacher CV. Purohit V, et al. Biophys J. 2024 Mar 5;123(5):622-637. doi: 10.1016/j.bpj.2024.02.003. Epub 2024 Feb 6. Biophys J. 2024. PMID: 38327055 - Structure of the complex between calmodulin and a functional construct of eukaryotic elongation factor 2 kinase bound to an ATP-competitive inhibitor.
Piserchio A, Isiorho EA, Dalby KN, Ghose R. Piserchio A, et al. J Biol Chem. 2023 Jun;299(6):104813. doi: 10.1016/j.jbc.2023.104813. Epub 2023 May 11. J Biol Chem. 2023. PMID: 37172726 Free PMC article. - Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Lin W, Huber AD, Poudel S, Li Y, Seetharaman J, Miller DJ, Chen T. Lin W, et al. Proc Natl Acad Sci U S A. 2023 Mar 7;120(10):e2217804120. doi: 10.1073/pnas.2217804120. Epub 2023 Feb 27. Proc Natl Acad Sci U S A. 2023. PMID: 36848571 Free PMC article. - Celastrol suppresses colorectal cancer via covalent targeting peroxiredoxin 1.
Xu H, Zhao H, Ding C, Jiang D, Zhao Z, Li Y, Ding X, Gao J, Zhou H, Luo C, Chen G, Zhang A, Xu Y, Zhang H. Xu H, et al. Signal Transduct Target Ther. 2023 Feb 3;8(1):51. doi: 10.1038/s41392-022-01231-4. Signal Transduct Target Ther. 2023. PMID: 36732502 Free PMC article.
References
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. - PubMed
- Greer, D. S., Westbrook, J. D. & Bourne, P. E. (2002). Bioinformatics, 18, 1280–1281. - PubMed
- Dai, J., Liu, J., Deng, Y., Smith, T. M. & Lu, M. (2004). Cell, 116, 649–659. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources