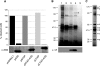Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules - PubMed (original) (raw)
Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules
Sarah Gallois-Montbrun et al. J Virol. 2007 Mar.
Abstract
Members of the APOBEC (apolipoprotein B mRNA-editing enzyme catalytic polypeptide 1-like) family of cytidine deaminases inhibit host cell genome invasion by exogenous retroviruses and endogenous retrotransposons. Because these enzymes can edit DNA or RNA and potentially mutate cellular targets, their activities are presumably regulated; for instance, APOBEC3G (A3G) recruitment into high-molecular-weight ribonucleoprotein (RNP) complexes has been shown to suppress its enzymatic activity. We used tandem affinity purification together with mass spectrometry (MS) to identify protein components within A3G-containing RNPs. We report that numerous cellular RNA-binding proteins with diverse roles in RNA function, metabolism, and fate determination are present in A3G RNPs but that most interactions with A3G are mediated via binding to shared RNAs. Confocal microscopy demonstrated that substantial quantities of A3G localize to cytoplasmic microdomains that are known as P bodies and stress granules (SGs) and are established sites of RNA storage and metabolism. Indeed, subjecting cells to stress induces the rapid redistribution of A3G and a number of P-body proteins to SGs. Among these proteins are Argonaute 1 (Ago1) and Argonaute 2 (Ago2), factors that are important for RNA silencing and whose interactions with A3G are resistant to RNase treatment. Together, these findings reveal that A3G associates with RNPs that are found throughout the cytosol as well as in discrete microdomains. We also speculate that the interplay between A3G, RNA-silencing pathways, and cellular sites of RNA metabolism may contribute to A3G's role as an inhibitor of retroelement mobility and as a possible regulator of cellular RNA function.
Figures
FIG. 1.
Purification of proteins associated with A3G RNP complexes. (A) Effect of A3G, NTAP-A3G, and CTAP-A3G on HIV-1 infectivity. GFP reporter viruses were produced in 293T cells in the absence of Vif and in the presence of either A3G, NTAP-A3G, CTAP-A3G, or a control vector. Normalized viral stocks were used to challenge HeLa cells, and infectivity was scored by assessing GFP expression using flow cytometry. Values are presented as percent infectivity relative to that for virus produced in the absence of A3G. Data are the means of two independent experiments. The lower panel shows an immunoblot analysis confirming comparable levels of expression of A3G and TAP-tagged A3G in virus-producing 293T cells. (B) TAP tag purification of A3G RNP complexes from 293T cells. Cultured monolayers were transfected with pNTAP-A3G alone (lanes 1 and 2), with pNTAP-A3G and pcVIF (lanes 3 and 4), or with the empty vector pNTAP (lane 5). Cell lysates were prepared and treated with RNase A for 1 h at room temperature (lanes 2 and 4) or left untreated (lanes 1 and 3). NTAP-A3G complexes were purified in two steps using streptavidin-binding peptide-coated beads and then calmodulin-binding peptide-coated beads. Final samples were resolved using 10% SDS-polyacrylamide gels, and the bands were visualized by silver staining. The band indicated by an arrow corresponds to full-length NTAP-A3G, and the band indicated by a dot was identified by MS as cullin5 (lane 4). The lower panel shows an immunoblot analysis confirming the presence of Vif in NTAP-A3G complexes. (C) TAP tag purification of A3G RNP complexes from CEM-SS cells stably expressing the NTAP-A3G protein. The methods are as described for panel B. The prominent bands were excised (arrows 1 to 8), and the proteins were then identified by mass spectrometry (refer to Table 1).
FIG. 2.
Coimmunoprecipitation of cellular proteins with A3G. Transfected 293T cells coexpressing A3G-HA (or empty vector as indicated) and tagged versions of cellular proteins (except LRP130) identified in the NTAP-A3G RNP complexes or Vif were lysed and immunoprecipitated with anti-HA or anti-myc antibodies and protein A or G linked to agarose. When indicated, the immunoprecipitates were treated with an RNase mixture (w/o, without RNase treatment). Samples were analyzed by immunoblotting using the indicated primary antibodies. In the La analyses, a background band of mobility slightly faster than that of tagged La was consistently seen for immunoprecipitated samples. For YB-1, the coimmunoprecipitation was reversed and A3G was detected on the blot. Bands indicated by asterisks correspond to immunoglobulin heavy or light chains.
FIG. 3.
A3G is localized in the cytoplasm and is enriched in discrete cytoplasmic foci. PBMCs (A), H9 cells (B), CEM-SS cells stably expressing the NTAP-A3G protein (C), or HeLa cells transfected with pA3G (D) were fixed and probed with rabbit anti-A3G antibodies. The cells were then stained with a rabbit-specific secondary antibody directly conjugated to Alexa Fluor 594 dye. Cell nuclei were stained with DAPI dilactate, and the samples examined by confocal microscopy. Scale bars = 10 μm.
FIG. 4.
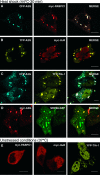
A3G colocalizes with P bodies in the cytoplasm. HeLa cells coexpressing YFP-A3G, CFP-A3G, or myc-A3G (left panels) and the indicated P-body markers myc-Mov10 (A), myc-Ago1 (B), myc-Ago2 (C), RFP-rck/p54 (D), mRFP-Dcp1a (E), GFP-GW182 (F), and myc-YB-1 (G) (middle panels) were fixed and stained with the 9E10 antibody and a mouse-specific secondary antibody conjugated to Alexa Fluor 594 dye. Samples were viewed by confocal microscopy, and merged views are displayed on the right. Scale bars = 10 μm.
FIG. 5.
A3G localizes to stress granules after heat shock. HeLa cells coexpressing CFP-A3G, YFP-A3G, or myc-A3G (left panels) and the indicated SG markers myc-PABPC1 (A), myc-HuR (B), YFP-TIA-1 (C), or SRP68-GFP (D) were incubated at 44°C for 30 min to induce SG formation. (E) Patterns of localization of myc-PABPC1, myc-HuR, and YFP-TIA-1 under normal cell growth conditions. Fixed cells were stained with the 9E10 antibody (A, B, D, and the first two panels of E), processed as for Fig. 4 and viewed by confocal microscopy. Merged views are displayed to the right. Scale bars = 10 μm.
FIG. 6.
Some P-body proteins colocalize to stress granules with A3G after heat shock. HeLa cells coexpressing CFP-A3G, YFP-TIA-1 (to define SGs), and the P-body markers myc-Mov10 (A), myc-Ago2 (B), myc-YB1 (C), and mRFP-Dcp1a (D) were stressed and analyzed as for Fig. 5. Merged views are displayed to the right. Scale bars = 10 μm.
FIG. 7.
Interactions of A3G with P-body proteins. Transfected 293T cells coexpressing A3G-HA (or empty vector) and tagged versions of P-body markers were lysed, immunoprecipitated for A3G-HA with an anti-HA antibody (A) or for Ago1/Ago2-myc with an anti-myc antibody (B), and subjected to RNase digestion as described for Fig. 2. Samples were analyzed by immunoblotting using the indicated primary antibodies. w/o, without.
Similar articles
- High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition.
Chiu YL, Witkowska HE, Hall SC, Santiago M, Soros VB, Esnault C, Heidmann T, Greene WC. Chiu YL, et al. Proc Natl Acad Sci U S A. 2006 Oct 17;103(42):15588-93. doi: 10.1073/pnas.0604524103. Epub 2006 Oct 9. Proc Natl Acad Sci U S A. 2006. PMID: 17030807 Free PMC article. - The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules.
Kozak SL, Marin M, Rose KM, Bystrom C, Kabat D. Kozak SL, et al. J Biol Chem. 2006 Sep 29;281(39):29105-19. doi: 10.1074/jbc.M601901200. Epub 2006 Aug 3. J Biol Chem. 2006. PMID: 16887808 - Comparison of cellular ribonucleoprotein complexes associated with the APOBEC3F and APOBEC3G antiviral proteins.
Gallois-Montbrun S, Holmes RK, Swanson CM, Fernández-Ocaña M, Byers HL, Ward MA, Malim MH. Gallois-Montbrun S, et al. J Virol. 2008 Jun;82(11):5636-42. doi: 10.1128/JVI.00287-08. Epub 2008 Mar 26. J Virol. 2008. PMID: 18367521 Free PMC article. - Multifaceted antiviral actions of APOBEC3 cytidine deaminases.
Chiu YL, Greene WC. Chiu YL, et al. Trends Immunol. 2006 Jun;27(6):291-7. doi: 10.1016/j.it.2006.04.003. Epub 2006 May 4. Trends Immunol. 2006. PMID: 16678488 Review. - Regulation of Antiviral Innate Immunity Through APOBEC Ribonucleoprotein Complexes.
Salter JD, Polevoda B, Bennett RP, Smith HC. Salter JD, et al. Subcell Biochem. 2019;93:193-219. doi: 10.1007/978-3-030-28151-9_6. Subcell Biochem. 2019. PMID: 31939152 Review.
Cited by
- Antiviral Defence Mechanisms during Early Mammalian Development.
Mueller F, Witteveldt J, Macias S. Mueller F, et al. Viruses. 2024 Jan 24;16(2):173. doi: 10.3390/v16020173. Viruses. 2024. PMID: 38399949 Free PMC article. Review. - Restricting retrotransposons: a review.
Goodier JL. Goodier JL. Mob DNA. 2016 Aug 11;7:16. doi: 10.1186/s13100-016-0070-z. eCollection 2016. Mob DNA. 2016. PMID: 27525044 Free PMC article. Review. - Effects of Moloney Leukemia Virus 10 Protein on Hepatitis B Virus Infection and Viral Replication.
Puray-Chavez MN, Farghali MH, Yapo V, Huber AD, Liu D, Ndongwe TP, Casey MC, Laughlin TG, Hannink M, Tedbury PR, Sarafianos SG. Puray-Chavez MN, et al. Viruses. 2019 Jul 17;11(7):651. doi: 10.3390/v11070651. Viruses. 2019. PMID: 31319455 Free PMC article. - Targeting APOBEC3A to the viral nucleoprotein complex confers antiviral activity.
Goila-Gaur R, Khan MA, Miyagi E, Kao S, Strebel K. Goila-Gaur R, et al. Retrovirology. 2007 Aug 29;4:61. doi: 10.1186/1742-4690-4-61. Retrovirology. 2007. PMID: 17727729 Free PMC article. - Non-long terminal repeat (non-LTR) retrotransposons: mechanisms, recent developments, and unanswered questions.
Han JS. Han JS. Mob DNA. 2010 May 12;1(1):15. doi: 10.1186/1759-8753-1-15. Mob DNA. 2010. PMID: 20462415 Free PMC article.
References
- Afonina, E., M. Neumann, and G. N. P. Pavlakis. 1997. Preferential binding of poly(A)-binding protein 1 to an inhibitory RNA element in the human immunodeficiency virus type 1 gag mRNA. J. Biol. Chem. 272:2307-2311. - PubMed
- Alce, T. M., and W. Popik. 2004. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 279:34083-34086. - PubMed
- Anant, S., and N. O. Davidson. 2003. Hydrolytic nucleoside and nucleotide deamination, and genetic instability: a possible link between RNA-editing enzymes and cancer? Trends Mol. Med. 9:147-152. - PubMed
- Anderson, P., and N. Kedersha. 2002. Stressful initiations. J. Cell Sci. 115:3227-3234. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources