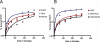Fragile X related protein 1 isoforms differentially modulate the affinity of fragile X mental retardation protein for G-quartet RNA structure - PubMed (original) (raw)
Fragile X related protein 1 isoforms differentially modulate the affinity of fragile X mental retardation protein for G-quartet RNA structure
Elias Bechara et al. Nucleic Acids Res. 2007.
Abstract
Fragile X syndrome, the most frequent form of inherited mental retardation, is due to the absence of expression of the Fragile X Mental Retardation Protein (FMRP), an RNA binding protein with high specificity for G-quartet RNA structure. FMRP is involved in several steps of mRNA metabolism: nucleocytoplasmic trafficking, translational control and transport along dendrites in neurons. Fragile X Related Protein 1 (FXR1P), a homologue and interactor of FMRP, has been postulated to have a function similar to FMRP, leading to the hypothesis that it can compensate for the absence of FMRP in Fragile X patients. Here we analyze the ability of three isoforms of FXR1P, expressed in different tissues, to bind G-quartet RNA structure specifically. Only the longest FXR1P isoform was found to be able to bind specifically the G-quartet RNA, albeit with a lower affinity as compared to FMRP, whereas the other two isoforms negatively regulate the affinity of FMRP for G-quartet RNA. This result is important to decipher the molecular basis of fragile X syndrome, through the understanding of FMRP action in the context of its multimolecular complex in different tissues. In addition, we show that the action of FXR1P is synergistic rather than compensatory for FMRP function.
Figures
Figure 1
The FXR1P isoforms. (A) Schematic representation of the C-terminal region of the three FXR1P isoforms analyzed: Isoe 84 kDa, Isod 78 kDa, Isoa 70 kDa. In the upper part of the figure the alternatively spliced sequences are indicated. A (+) under each amino acid indicates the predicted ability of the sequence to bind RNA accordingly to the algorithm described by Terribilini and coworkers (28). (B) Production of recombinant proteins. Equal amounts of Hist-FMRP, His-FXR1P-Isoe, Isod, Isoa and GST-MSP58 were loaded on a 10% SDS–PAGE gel and revealed by Coomassie blue staining.
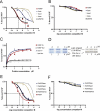
Figure 2
RNA binding properties of FMRP and FXR1P isoforms. (A) Filter binding assay using FMRP, FXR1P-Isoe, Isod, Isoa and MSP58. The RNA probe used is 32P-labeled N19 RNA, and competition was performed using the same unlabeled RNA. (B) The same experiment was repeated using as competitor the N8 RNA sequence, not containaing any G-quartet forming structure. (C) Filter binding assay was repeated with an increasing amount of FMRP and Isoe in the presence of Na+ or K+. (D) GST-pull down was performed as described in Materials and Methods. On the right part of (D), proteins used in GST-pull down assay were revealed by immunoblot. FMRP was detected by monoclonal 1C3 antibody, FXR1P by the monoclonal 3FX antibody. Lane 1: 4 μM GST-FMRP complexed with 1 μM His-FXR1P, Lane 2: 4 μM GST-FMRP complexed with 2 μM His-FXR1P, Lane 3: 4 μM GST-FMRP complexed with 4 μM His-FXR1P. On the left part of (D), proteins used in GST-pull down experiment were revealed by Coomassie stained gel. (E) Competition assay to determine the _K_d at the equilibrium state binding FMRP, the heterodimers FMRP/Isoe, FMRP/Isod, FMRP/Isoa and the complex FMRP/MSP58, with the 32P-labeled N19 probe and competed with unlabeled N19. (F) The same experiment described in (E) was repeated using the N8 RNA as unlabeled competitor.
Figure 3
Association rate of FMRP, Isoe and MSP58. (A) Each protein was mixed with 32P-labeled N19 RNA probe for a time lapse of 10, 30, 60, 120, 180, 240 and 300 min and then each reaction was filtered and the amount of retained radioactivity evaluated. (B) The same experiment described in (A) was repeated with the complex FMRP/MSP58 and FMRP/Isoe as indicated in the figure.
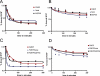
Figure 4
Dissociation rate of FMRP, Isoe, MSP58 and the two complexes FMRP/Isoe and FMRP/MSP58. (A) Each protein was mixed with 32P-labeled N19 RNA probe for 10 min on ice and then an equal amount of unlabeled N19 RNA was added as competitor to each reaction, which was then filtered after a precise time lapse of 10, 30, 60, 120, 180, 240 and 300 min. The radioactivity retained on the filter was evaluated. (B) The same experiment described in (A) was repeated using unlabeled N8 RNA as competitor. (C) The same experiment described in (A) was performed using the protein complexes FMRP/Isoe and FMRP/MSP58. (D) The same experiment described in (C) was performed using the cold N8 RNA as competitor.
Similar articles
- Discrimination of common and unique RNA-binding activities among Fragile X mental retardation protein paralogs.
Darnell JC, Fraser CE, Mostovetsky O, Darnell RB. Darnell JC, et al. Hum Mol Genet. 2009 Sep 1;18(17):3164-77. doi: 10.1093/hmg/ddp255. Epub 2009 Jun 1. Hum Mol Genet. 2009. PMID: 19487368 Free PMC article. - Thermodynamics of the fragile X mental retardation protein RGG box interactions with G quartet forming RNA.
Zanotti KJ, Lackey PE, Evans GL, Mihailescu MR. Zanotti KJ, et al. Biochemistry. 2006 Jul 11;45(27):8319-30. doi: 10.1021/bi060209a. Biochemistry. 2006. PMID: 16819831 - Analysis of the Fragile X mental retardation protein isoforms 1, 2 and 3 interactions with the G-quadruplex forming semaphorin 3F mRNA.
Evans TL, Blice-Baum AC, Mihailescu MR. Evans TL, et al. Mol Biosyst. 2012 Feb;8(2):642-9. doi: 10.1039/c1mb05322a. Epub 2011 Dec 1. Mol Biosyst. 2012. PMID: 22134704 - Biology of the fragile X mental retardation protein, an RNA-binding protein.
Khandjian EW. Khandjian EW. Biochem Cell Biol. 1999;77(4):331-42. Biochem Cell Biol. 1999. PMID: 10546896 Review. - The Fragile X mental retardation protein.
Bardoni B, Schenck A, Mandel JL. Bardoni B, et al. Brain Res Bull. 2001 Oct-Nov 1;56(3-4):375-82. doi: 10.1016/s0361-9230(01)00647-5. Brain Res Bull. 2001. PMID: 11719275 Review.
Cited by
- FXR1 associates with and degrades PDZK1IP1 and ATOH8 mRNAs and promotes esophageal cancer progression.
Khan FA, Fouad D, Ataya FS, Fang N, Dong J, Ji S. Khan FA, et al. Biol Direct. 2024 Nov 7;19(1):104. doi: 10.1186/s13062-024-00553-3. Biol Direct. 2024. PMID: 39511680 Free PMC article. - PRMT5-mediated arginine methylation of FXR1 is essential for RNA binding in cancer cells.
Vijayakumar A, Majumder M, Yin S, Brobbey C, Karam J, Howley B, Howe PH, Berto S, Madan LK, Gan W, Palanisamy V. Vijayakumar A, et al. Nucleic Acids Res. 2024 Jul 8;52(12):7225-7244. doi: 10.1093/nar/gkae319. Nucleic Acids Res. 2024. PMID: 38709899 Free PMC article. - FXR1 regulates vascular smooth muscle cell cytoskeleton, VSMC contractility, and blood pressure by multiple mechanisms.
St Paul A, Corbett C, Peluzzo A, Kelemen S, Okune R, Haines DS, Preston K, Eguchi S, Autieri MV. St Paul A, et al. Cell Rep. 2023 Apr 25;42(4):112381. doi: 10.1016/j.celrep.2023.112381. Epub 2023 Apr 11. Cell Rep. 2023. PMID: 37043351 Free PMC article. - FMRP, FXR1 protein and Dlg4 mRNA, which are associated with fragile X syndrome, are involved in the ubiquitin-proteasome system.
Shimizu H, Hohjoh H. Shimizu H, et al. Sci Rep. 2023 Feb 2;13(1):1956. doi: 10.1038/s41598-023-29152-4. Sci Rep. 2023. PMID: 36732356 Free PMC article. - FMRP, a multifunctional RNA-binding protein in quest of a new identity.
Khandjian EW, Robert C, Davidovic L. Khandjian EW, et al. Front Genet. 2022 Aug 10;13:976480. doi: 10.3389/fgene.2022.976480. eCollection 2022. Front Genet. 2022. PMID: 36035132 Free PMC article. No abstract available.
References
- Bardoni B., Mandel J.L., Fisch G.S. FMR1 gene and fragile X syndrome. Am. J. Med. Genet. 2000;97:153–163. - PubMed
- Bardoni B., Davidovic L., Bensaid M., Khandjian E.W. The fragile X syndrome: exploring its molecular basis and seeking a treatment. Expert Rev. Mol. Med. 2006;8:1–16. - PubMed
- Bakker C.E., Verheij C., Willemsen R., Vanderhelm R., Oerlemans F., Vermey M., Bygrave A., Hoogeveen A.T., Oostra B., Reyners E., et al. Fmr1 knockout mice: a model to study fragile X mental retardation. Cell. 1994;78:23–33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials